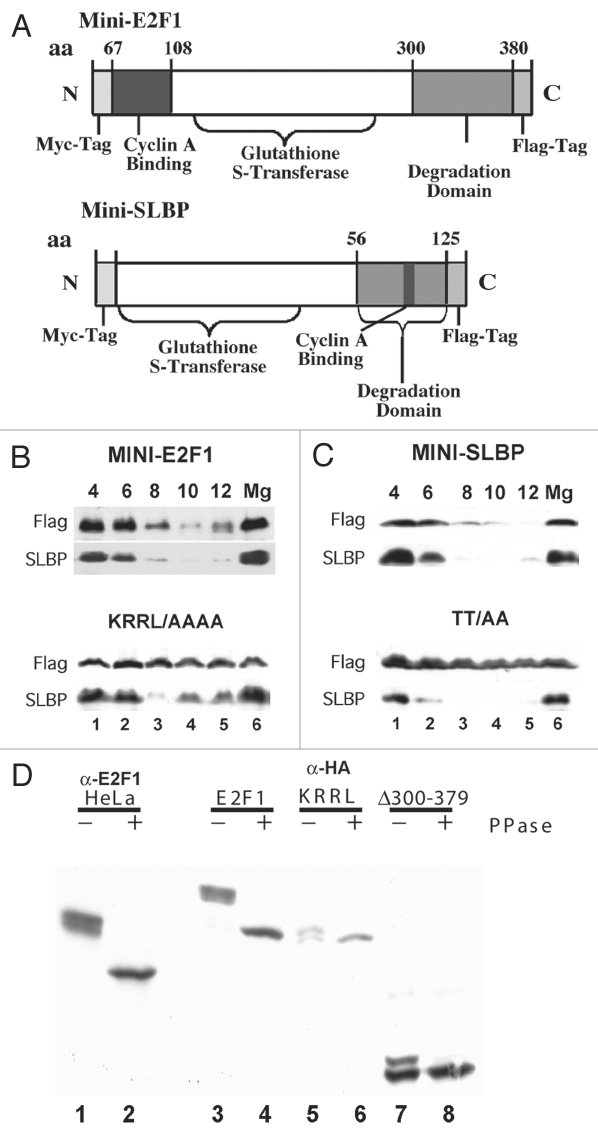
Figure 3.
Phosphorylation of E2F1 requires a cyclin binding site for E2F1 degradation. (A) The indicated minigenes for SLBP5 and E2F1 are shown. Amino acids 56–125 of SLBP are sufficient to direct degradation of the protein. The E2F minigene contains the two fragments of E2F1 identified in Figure 2, 67–108 and 300–380 separated by GST. Point mutants of the core of the cyclin binding site of E2F1 and in the TT which is the target of phosphorylation of SLBP were also constructed in the context of the minigene. (B and C) Cells stably expressing the E2F1 minigene and mutant minigene (B) or the SLBP minigene or mutant minigene (C) were synchronized by double-thymidine block, released into S phase. Parallel cultures expressing the SLBP and E2F minigenes were synchronized and lysates were prepared at the indicated times after release and analyzed by western blotting for the FLAG-tag on the minigene or SLBP antibody to detect endogenous SLBP. (D) Cells expressing HA-tagged wild-type E2F1, the cyclin binding site mutant in the wild-type protein, of the 300–379 deletion (lanes 3–8) were synchronized by double-thymidine block and lysate prepared 4 hrs after release into S phase. A portion of the lysate was treated with lambda protein phosphatase. Equal amounts of the treated and untreated lysates were analyzed by western blotting using the anti-HA antibody after resolution of long polyacrylamide gels for an extended amount of time to resolve phosphorylated forms. The endogenous E2F1 was analyzed in lanes 1 and 2 using the E2F1 antibody. Note the difference in mobility of the endogenous E2F1 and the tagged-E2F1 is due to the HA-tag.