Abstract
In Eukarya, the packaging of DNA into chromatin provides a barrier that allows for regulation of access to the genome. Chromatin is refractory to processes acting on DNA. ATP-dependent chromatin remodeling machines and histone-modifying complexes can overcome this barrier (or strengthen it in silencing processes). Both components of chromatin (DNA and histones) are subject to postsynthetic covalent modifications, including methylation of lysines (the focus of this chapter). These lysine marks are generated by a host of histone lysine methyltransferases (writers) and can be removed by histone lysine demethylases (erasers). Importantly, epigenetic modifications impact chromatin structure directly or can be read by effector regulatory modules. Here, we summarize current knowledge on structural and functional properties of various histone lysine methyltransfereases and demethylases, with emphasis on their importance as druggable targets.
1 Introduction
Unlike lysine acetylation, methylation of lysines does not alter the effective charge, but the hydrophobic and steric properties. The degree of lysine methylation can be mono-, di-, or tri-methylated depending on the specific functional properties of the associated methyltransferase [1–3]. These different lysine methylation marks serve as the binding site for different effector proteins with cognate recognition domains specific to different methylated lysine residues. For example, plant homeodomain (PHD) of bromodomain-PHD-transcription-factor (BPTF) binds tri- or di-methylated lysine 4 of histone H3 (H3K4me3/me2) and recruits the nucleosome remodeling factor (NURF) complex to the target gene leading to gene activation [4, 5]. In an opposite mechanism, the chromodomain of heterochromatin protein 1 (HP1) binds tri-methylated lysine 9 of histone H3 (H3K9me3) mark, which initiates heterochromatin formation and gene silencing [6, 7].
Recent evidences have indicated that specific recognition domains, either present in a protein complex or in the same polypeptide, combinatorially recognize different histone modifications through a crosstalk mechanism leading to the propagation of active or repressive state of the chromatin. One such example includes the polycomb repressive complex 2 (PRC2) in maintaining and propagating repressive tri-methylated lysine 27 of histone H3 (H3K27me3) through allosteric interaction between EZH2 and EED subunits [8]. Similar examples also include histone lysine methylating enzymes like mammalian G9a and G9a-like protein (GLP) (for H3K9me2/me1) and yeast Clr4 (for H3K9me3), containing both a catalytic SET domain and methyl-lysine recognition module (ankyrin repeats or chromodomain) within the same polypeptide [9, 10]. Therefore, methylation of specific lysines on histones regulates the recruitment of various downstream DNA processing proteins onto the chromatin, which in turn regulate a multitude of biological processes including heterochromatin formation, X-chromosome inactivation, DNA methylation, and gene silencing [11, 12].
The extensively studied histone lysine methylation marks include lysines 4, 9, 27, 36, and 79 of histone H3 and lysine 20 of histone H4. In general, H3K4, H3K36, and H3K79 methylation have been associated with transcriptionally active euchromatin, whereas H3K9, H3K27, and H4K20 methylations are associated with transcriptional inactive heterochromatin [2, 11]. Aberrant methylation of histone lysines has been implicated in various disease etiologies including cancer and X-linked mental retardation [3, 12–15]. Therefore, a proper understanding of the structural and functional regulations of the enzymes responsible for reversible modifications of histone lysines is of immense importance in developing future therapeutics for many of these diseases. Following is a summary of our understanding on the structural properties of known enzymes responsible for catalyzing specific lysine methylation and enzymes responsible for selective removal of these methylation marks.
2 Histone Lysine (K) Methyltransferases (HKMTs)
With the exception of Dot1 [16–18], all known HKMTs contain an evolutionarily conserved SET domain comprised of 130 amino acids [19–23]. The SET domain was first identified as a shared sequence motif in three Drosophila proteins, suppressor of variegation [Su(var)3–9], enhancer of zeste [E(z)], and homeobox gene regulator trithorax [Trx] [24]. Mammalian homologues of Drosophila Su(var) 3–9 protein, SUV39H1 in human and Suv39h in mouse, were the first characterized HKMTs involved in H3K9 methylation [24]. Since then, more than 50 SET domain-containing proteins with proven or predicted enzymatic role in carrying out lysine methylation on histone tail have been identified in human [19, 25].
With a few exceptions (e.g., Set8), the majority of the SET-containing HKMTs contain at least one additional protein module in their protein sequence. Based on the sequence homology within and around the catalytic SET domain, as well as based on other protein modules and their architectures, SET-containing HKMTs are grouped into six different subfamilies: SET1, SET2, SUV39, EZH, SMYD, and PRDM [19, 20, 25]. A number of SET-containing HKMTs, however, do not fall into the above six subfamilies, due to lacking sequences (and conservation) flanking their SET domains. Examples of such proteins include Set8/PR_Set7 (mono-methylates H4K20), SUV4-20H1 and SUV4-20H2 (di- and tri-methylates H4K20), Set7/9 (mono-methylates H3K4 and many other nonhistone substrates), as well as MLL5, SetD5 (KIAA1757), and SetD6 (FLJ21148) with currently unknown role in histone lysine methylation.
3 Structures of SET Domains
Structures of many SET domains from different subfamilies have been solved in various combinations with bound substrate peptide and methyl donor (S-adenosyl-l-methionine, AdoMet) or reaction product (S-adenosyl-l-homocysteine, AdoHcy) (Table 1). Representative structures of the SET-domain are displayed in Fig. 1. The SET domain adopts a unique structural design formed by a series of β-strands folded into three sheets surrounding a knot-like structure (Fig. 1). The knot-like structure is formed by the C-terminal segment of the SET domain, which passes through a loop formed by the preceding stretch of sequences. Formation of this knot-like structure brings two conserved sequence motifs of the SET domain, consisting of RFINHxCxPN and ELx(F/Y)DY, in close proximity to the AdoMet-binding region and peptide-binding channel (Fig. 2a). Interestingly, biochemical studies performed with F/Y mutants of the conserved ELx(F/Y)DY motif in DIM-5 (F281Y), G9a (F1205Y), Set8 (Y334F), Set7/9 (Y305F), and Set1 (Y1052F) suggest that the F/Y switch regulates the product specificity (mono-, di-, or tri-methylation) of SET-containing HKMTs [26–29].
Table 1.
List of HKMTs with known structures (PDB ID)
| Position | HKMT | PDB ID |
|---|---|---|
| H3K4 | MLL1 | 2W5Y, 2W5Z |
| SET7/9 (including nonhistone substrates) | 3CBO, 3CBM, 3CBP, 2F69, 1XQH, 1O9S, 1N6C, 1N6A, 1H3I, 1MUF, 1MT6 | |
| H3K9 | SUV39H2 | 2R3A |
| G9a (EHMT2) | 2O8J, 3K5K | |
| GLP (EHMT1) | 2RFI, 3FPD, 3HNA, 2IGQ, | |
| RIZ1 (PRDM2) | 2JV0, 2QPW | |
| DIM-5 | 1PEG, 1ML9 | |
| Clr4 | 1MVX, 1MVH | |
| H3K36 | SET2 | 3H6L |
| H3K79 | DOT1L | 1NW3, 1U2Z |
| H4K20 | PR-SET7 (SET8) | 3F9W, 3F9X, 3F9Y, 3F9Z, 2BQZ, 1ZKK |
| Others | PRDM10 | 3IHX |
| PRDM12 | 3EP0 | |
| PRDM1 | 3DAL | |
| SETMER | 3BO5 |
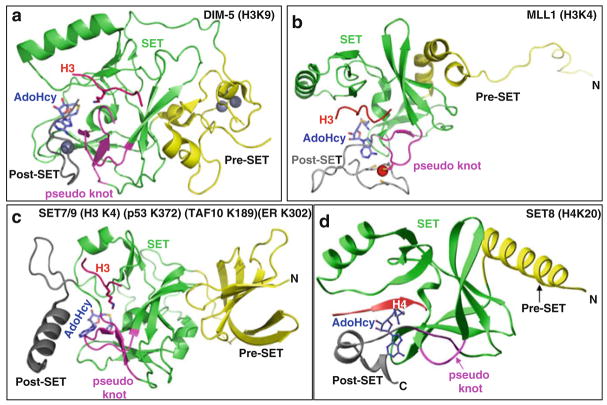
Fig. 1.
Examples of SET domain structures. Ribbon diagram of (a) Neurospora DIM-5 [26], (b) human MLL1 [102], (c) human SET7/9 [31], and (d) human SET8 [103] (or PR-SET7 [104])
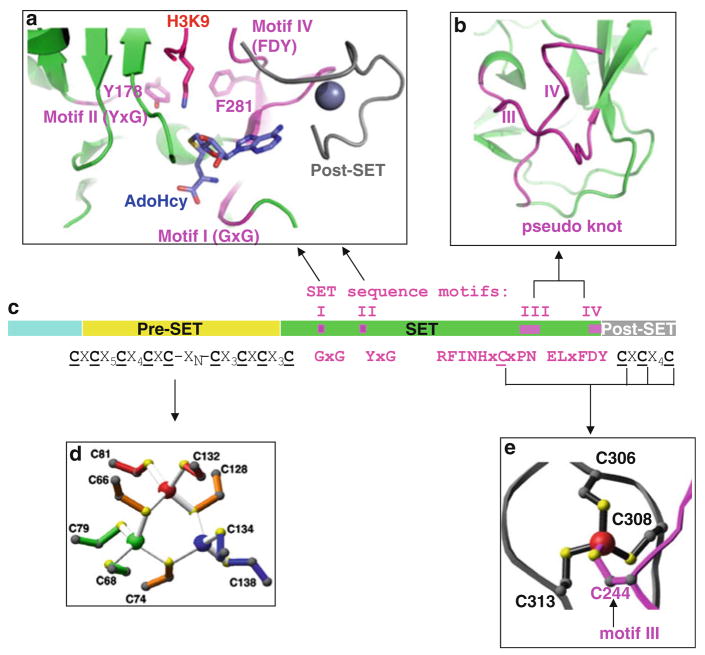
Fig. 2.
Structural features of Neurospora DIM-5 [26, 30]. (a) Ribbon diagram of the pseudo knot formed by motifs III and IV. (b) DIM-5 contains four segments: a weakly conserved amino-terminal region, a pre-SET domain containing nine invariant cysteines, the SET region containing four signature motifs, and the post-SET domain containing three invariant cysteines. (c) Illustration of pre-SET Zn3Cys9 triangular zinc cluster and (d) post-SET zinc center
4 Structural Properties of Pre-SET and Post-SET Modules
Available crystal structures of the SUV39 subfamily (DIM-5, Clr4, GLP/EHMT1, G9a/EHMT2, and SUV39H2 – all H3K9 HKMTs) show the presence of two closely packed cysteine rich-modules in the pre-SET and post-SET (before and after the SET domain) (Fig. 1a). These two modules are important in maintaining structural stability (pre-SET) and forming part of the active site lysine channel (post-SET) [26, 30]. The pre-SET module of SUV39 subfamily contains nine conserved cysteines (Fig. 2b), which coordinate three Zn2+ atoms in a triangular geometry (Fig. 2c). The post-SET module of SUV39 as well as Set1 (Fig. 1b) and Set2 subfamilies contains three conserved cysteines, which along with a cysteine from the conserved RFINHxCxPN motif of the SET domain tetrahedrally coordinate one Zn2+ atom near the active site (Fig. 2d). Binding of this Zn2+ at the active site is essential for the activity of SUV39 subfamily and therefore is a promising site for drug targeting [26].
The pre-SET and post-SET sequences in Set7/9 do not contain any cysteine-rich region. Instead, pre-SET in Set7/9 is occupied with a β-sheet structure comprised of 12 antiparallel β-strands, while the post-SET is occupied with a small α-helix [31] (Fig. 1c). Packing of the post-SET helix into the catalytic SET domain is important to form the substrate-binding groove in SET7/9. Similar variations in the sequences flanking the SET domain have also been observed in other subfamilies of HKMTs and suggest a convergent evolution of SET-containing HKMTs. This variation may also explain the differences in substrate specificities among the SET-containing HKMTs.
Structural and biochemical studies suggested that consensus substrate recognition sequences for G9a and Set7/9 contain only two to three residues: RK (G9a) and (R/K) (S/T)K (Set7/9) [32, 33]. The short recognition sequences enable these two enzymes to methylate many nonhistone substrates, including Set7/9-mediated methylation of p53 [34], components of the TBP complex, TAF10 [35] and TAF7 [33], estrogen receptor α [36], DNA methyltransferase 1 [37], and G9a-mediated methylation of chromodomain Y-like protein (CDYL1) and widely interspaced zinc finger motifs protein (WIZ) [32], CCAAT/enhancer-binding protein-β(C/EBPβ) [38], as well as G9a auto-methylation [39]. It appears that the dynamic lysine methylation of nonhistone proteins is a rapidly developing new field [40].
5 Structure of Inhibitor Bound G9a and GLP SET Domains
Methylation of H3K9 occurs in heterochromatin, which requires trimethylation of histone H3 at lysine 9 (H3K9me3) by Suv39h [41, 42], and in euchromatin, which requires mono- and di-methylation of H3K9 (H3K9me1/me2) mostly by G9a and GLP [43, 44]. H3K9me1/me2 are the only silencing marks that are lost when tumor suppressor genes, e.g., in colorectal cancer cells [45] and in breast cancer cells [46], are reactivated following treatment with 5-aza-2′-deoxycytidine, a DNA demethylation drug [47]. Thus, the enzymes that produce H3K9me1/me2 are appealing targets for inhibition.
A small molecule, BIX-01294 (a diazepin-quinazolin-amine derivative), was originally identified as a G9a inhibitor during a chemical library screen of small molecules [48]. The compound inhibits G9a and GLP activities (IC50 in low μM range) [48, 49] and reduces the methylation levels of H3K9 at several G9a target genes [48, 50]. BIX-01294 was used in combination with genetic factors to improve the efficiency of generation of induced pluripotent stem cells [51–53]. This is consistent with the observation that repressive H3K9 methylation by G9a is associated with the inactivation of Oct3/4, one of the four Yamanaka genetic factors required for included pluripotency [54], during differentiation [55].
BIX-01294 was crystallized with the catalytic SET domain of GLP in the presence of AdoHcy [49]. The inhibitor is bound in the acidic substrate peptide groove at the location where the histone H3 residues N-terminal to the target lysine lie. The inhibitor resembles the bound conformation of histone H3K4 to H3R8 and is positioned by residues specific for G9a and GLP through specific interactions. Most importantly, the inhibitor-bound SET domain structure provides avenues for improving the potency of the inhibitor. One of suggested improvements is by extending the branch of O7-methoxy-CH3 into the target lysine-binding channel, which should provide additional binding energy by increasing the surface area of binding [49]. Indeed, a recent report of chemical exploration of BIX-01294 identified a derivative (UNC0224) as a potent and selective G9a inhibitor [56]. UNC0224 contains an extended N-dimethylamino-propoxy arm occupying the target lysine-binding channel.
6 Histone Lysine Specific Demethylase (LSD1)
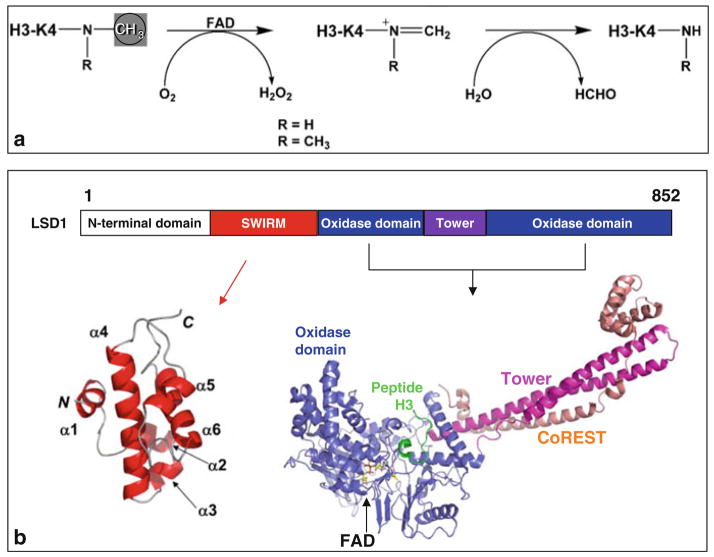
The discovery of lysine specific demethylase 1 (LSD1) [57] established that protein lysine methylation is a reversible posttranslational modification. LSD1 is a flavin-dependent amine oxidase, which demethylates H3K4me2/me1 [57], H3K9me2/me1 (in an androgen receptor-mediated pathway) [58], and p53 [59]. The closely related LSD2 demethylates H3K4me2/me1 [60] and has been linked with imprinting of the maternal genome [61]. Both LSD1 and LSD2 demethylate methyl-lysine by forming of an imine intermediate, which undergoes hydrolysis in aqueous buffer (Fig. 3a) to complete the demethylation process. Mechanistic requirements for a protonated amine in this demethylation pathway do not permit either LSD1 or LSD2 to demethylate trimethylated lysines [62].
Fig. 3.
Demethylation by oxidation. (a) Scheme of the demethylation reaction catalyzed by LSD1. (b) Schematic representation of human LSD1 domain organization. The oxidase domain contains an atypical insertion of the Tower domain not found in other oxidases. The solution NMR structure of the SIWRM domain of LSD1 is shown in red [65]. Crystal structure of LSD1 (residues 171–836 in blue)-CoREST (residues 308–440 in red ) in complex with H3 peptide (residues 1–16 in green), and the FAD cofactor is shown as a yellow ball-and-stick [71]
LSD1 is found in histone modification complexes that control cell-specific gene expression [57]. Within these complexes, REST (RE1-silencing transcription factor) corepressor CoREST enables LSD1 to demethylate nucleosomes [63, 64], while BHC80 (BRAF–HDAC complex) inhibits LSD1 activity [63]. The LSD1 polypeptide chain can be divided into several structural/functional regions (Fig. 3b): the N-terminal putative nuclear localization signal, followed by a SWIRM (Swi3p, Rsc8p, and Moira) domain [65] – found in several nucleosome-interacting proteins – and a monoamine oxidase domain – capable of demethylating lysines in a flavin-dependent manner [66]. Thus far, crystal structures of LSD1 alone [67, 68], LSD1 in complex with CoREST [69], and LSD1-CoREST in complex with H3 peptide [70, 71] have been determined. Using a 21-residue peptide bearing a methionine in place of target methyl-K4 – a 30-fold increase in binding affinity making the mutant peptide a strong inhibitor and an ideal candidate for structural work – Forneris et al. (2007) were able to resolve the first 16 residues of the H3 peptide, in perfect agreement with their previous biochemical data that LSD1 is active on peptide substrates longer than 16 amino acids [66]. This study is the first in which a long, structured histone tail has been visualized in histone-modifying enzymes and protein domains that recognize (decode) methyl-lysine signals. In comparison, a similar study of LSD1-histone peptide, using the approach of covalent tethering of peptide substrate to cofactor FAD, observed the first 7 residues (out of 21 residues used) of H3 peptide [70].
7 Jumonji-Containing Lysine Demethylases
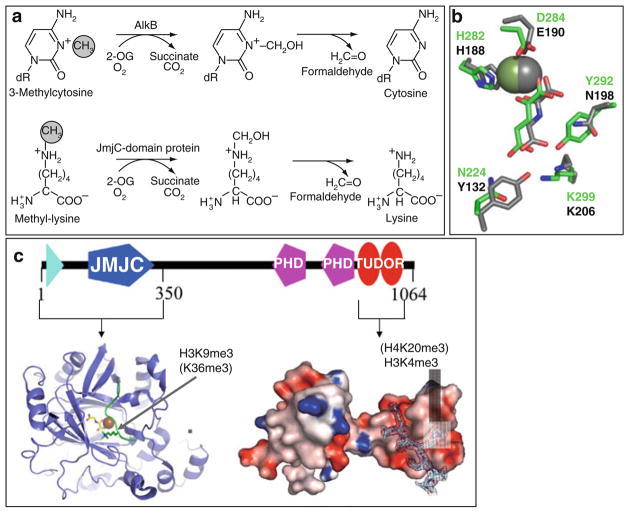
In search of enzymes capable of reversing methylated lysines, Trwick et al. [72] hypothesized that Jumonji domain containing Fe2+- and α-ketoglutarate-dependent dioxygenases can reverse lysine methylation via a similar mechanism as followed by bacterial AlkB family of DNA repair enzymes (Fig. 4a). This hypothesis was quickly verified with the discovery of JHDM1 as the Jumonji domain-containing histone demethylase 1 [73]. Jumonji-containing proteins are members of the cupin superfamily with functional roles in various biological processes including DNA/RNA repair through the demethylation of N-methylated nucleic acids (e.g., 3-methylcytosine, 1-methyladenine) [74, 75], hydroxylation of protein and lipid side chains [76], protein lysyl-5-hydroxylation [77], as well as recently characterized role in oxidizing 5-methylcytosine to 5-hydroxymethylcytosine [78]. Demethylation reactions catalyzed by Jumonji enzymes follow a hydroxylation pathway, which can demethylate mono-, di-, or tri-methylated lysines (Fig. 4a) [79, 80].
Fig. 4.
Demethylation by hydroxylation. (a) Mechanisms of demethylation of 3-methylcytosine by AlkB (top) and of methyl-lysine by Jumonji-domain proteins (bottom). (b) Coordinations of Fe2+ (sphere), α-ketoglutarate in JMJD2A (in gray), and KIAA1718 (in green). (c) Schematic representation of JMJD2A domain organization, including the structures of the N-terminal Jumonji (ribbons) [85] and the C-terminal double Tudor domain (surface representation) [87]
Currently, there are nearly 30 Jumonji-containing proteins identified in human proteome, 20 of which have known function in histone demethylation [2]. The majority of Jumonji-containing demethylases contains at least one additional structural domain in their sequence. Based on the phylogenetic relationships and domain architectures, these proteins are divided into seven subfamilies [2]. Additional structural motifs (other than the Jumonji domain) present in these proteins are thought to be important in substrate recognition or facilitating protein–protein interactions. For example, the H3K4 demethylase RBP2 contains a DNA-binding domain, the AT-rich interaction domain (ARID). ARID binds DNA sequence motif (CCGCCC) and is required for RBP2 demethylase activity in cells and that DNA recognition is essential to regulate transcription [81].
Thirteen crystal structures for the Jumonji domain of JMJD2A in various configurations are currently available (Table 2) [67, 82–85]. In addition, one structure is available for JMJD2D, two for JHDM1A [86], and two for PHF8 Jumonji domain (Table 2). Like in other cupin family members, the Jumonji domain adopts the conserved double-stranded-β-helix or jelly-roll structure formed by eight antiparallel β-strands, which harbors the Fe2+ (coordinated by two histones and one aspartate or glutamate) and α-ketoglutarate in a conserved coordination environment (Fig. 4b). The co-substrate α-ketoglutarate is coordinated to the Fe2+ center through C1-carboxylate and C2-keto group. The C5-carboxylate of α-ketoglutarate forms hydrogen-bonding interactions with Jumonji domain.
Table 2.
List of histone lysine demethylases with known structures (PDB ID)
| Position | HDM | PDB ID |
|---|---|---|
| H3K4 | LSD1 | 2IW5, 2HKO, 2V1D, 2UXN, 2UXX, 2DW4, 2Z3Y, 2EJR, 2Z5U |
| H3K9 | PHF8 | 3K3O, 3K3N |
| JMJD2A | 2VD7, 2Q8C, 2Q8D, 2Q8E, 2P5B, 2PXJ, 2OQ6, 2OQ7, 2OS2, 2OT7, 2OXO, 2GP3, 2GP5 | |
| JMJD2D | 3DXT | |
| H3K36 | JHDM1A (FBXL11) | 2YU1, 2YU2 |
8 JMJD2A
JMJD2A contains an N-terminal Jumonji domain and C-terminal PHD and Tudor domains (Fig. 4c). The JMJD2A Jumonji domain alone is capable of demethylating tri- and di-methylated H3K9 (H3K9me3/2) and H3K36 (H3K36me3/2), though with a very low turnover rate [84]. Structural studies revealed that the JMJD2A Jumonji domain predominantly recognizes the backbone of the histone peptides (unusual for a sequence-specific enzyme), allowing the enzyme to demethylate both H3K9me3/2 and H3K36me3/2 [83–85]. On the other hand, JMJD2A Tudor domain binds two different histone sequences (H3K4me3 and H4K20me3) via radically different approaches [87, 88]. The functional connection between the methyl mark reader and eraser in JMJD2A is not clear.
9 PHF8 and KIAA1718
PHF8 and KIAA1718 belong to a small family of Jumonji proteins with three members in mice and human (PHF2, PHF8, and KIAA1718) [2]. These proteins harbor two domains in the N-terminal half (Fig. 5a): a PHD domain that binds H3K4me3 and a Jumonji domain that demethylates H3K9me2, H3K27me2, as well as H3K36me2 [89]. However, the presence of H3K4me3 on the same peptide as H3K9me2 makes the doubly methylated peptide a significantly better substrate of PHF8 [90]. In contrast, the presence of H3K4me3 has the opposite effect in that it diminishes the H3K9me2 demethylase activity of KIAA1718 with no adverse effect on its H3K27me2 activity. Differences in substrate specificity between the two enzymes are explained by a bent conformation of PHF8, allowing each of its domains to engage their respective targets, and an extended conformation of KIAA1718, which prevents its access to H3K9me2 by its Jumonji domain when its PHD domain engages H3K4me3 (Fig. 5a). This study concludes that the structural linkage between the PHD domain binding to H3K4me3 and the placement of the catalytic Jumonji domains relative to this “on” epigenetic mark determines which repressive marks are removed in both demethylases. Taken together, we suggest that the PHF8 and KIAA1718 Jumonji domains on their own are promiscuous enzymes; it is the associated PHD domains and linker – a determinant for the relative positioning of the two domains – that are mainly responsible for substrate specificity.
Fig. 5.
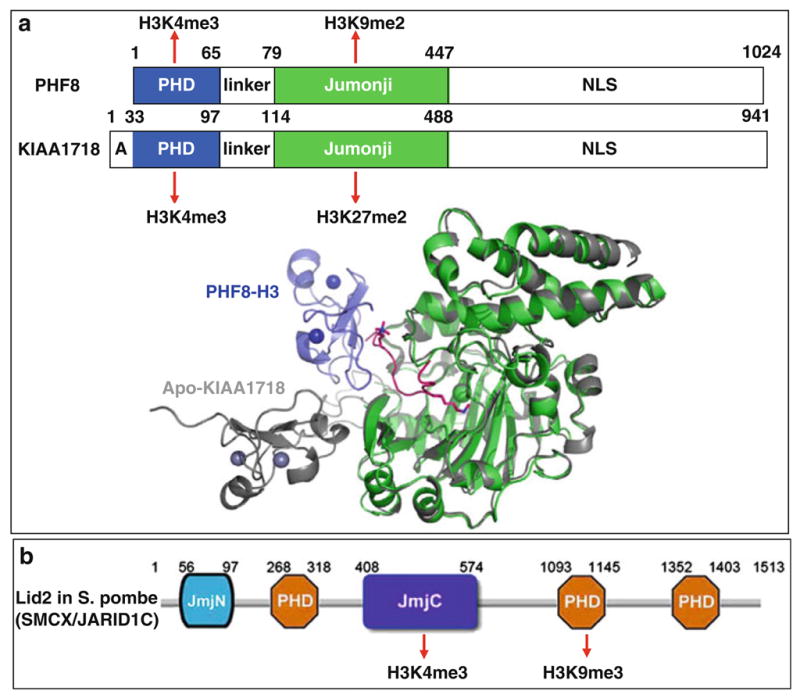
Crosstalk between Jumonji and PHD within the same polypeptide. (a) Schematic representations of PHF8 and KIAA1718. Superimposition of PHF8 (colored) and KIAA1718 (gray) in their respective Jumonji domains indicates that the PHF8 PHD domain adopts a bent conformation toward the Jumonji domain in the presence of H3 substrate binding, whereas the PHD and Jumonji domains of KIAA1718 adopt an extended conformation in its apo-structure [90]. (b) Schematic representation of Lid2 in S. pombe [93] (SMCX/JARID1C [92])
Using domain cooperativity to enhance an enzyme’s activity and its substrate specificity may be a general mechanism for Jumonji-containing protein lysine demethylases. For example, JHDM2A-mediated histone H3K9me1/2 demethylation requires a zinc finger N-terminal to the Jumonji domain for its enzymatic activity [91]. JARID Jumonji family proteins (including Lid2 in S. pombe) contain a Jumonji domain that demethylates H3K4me3 surrounded by several PHD domains and at least one of them binds H3K9me3 [92, 93] (Fig. 5b). Mutation or deletion of this PHD domain impairs the demethylase activity on H3K4me3 [92, 93]. We speculate that the ideal substrate for JARID family is H3 trimethylated at both K4 and K9, allowing the enzyme to remove any local activating methyl groups of H3K4me3 by the Jumonji in a repressing environment with H3K9me3 bound by the PHD (Fig. 5b). We further speculate that a similar situation might occur for JMJD2A where each of the two demethylase activities (H3K9me3/2 and H3K36me3/2) correlates with one of the methyl marks (H3K4me3 and H4K20me3) recognized by the Tudor domain (Fig. 4b).
10 Perspective
The histone code hypothesis suggests that multiple covalent histone modifications can be read combinatorially through effectors that are recruited to these marks and subsequently act on the local chromatin structure or transcriptional machinery via crosstalk among histone modifications [94–97]. Several histone-methylating enzymes contain components (domains) that both synthesize and bind a specific histone mark, such as mammalian G9a/GLP (for H3K9me1/me2) [9] and S. pombe Clr4 (for H3K9me3) [10]. They contain modules, within the same polypeptide, for both making (via the SET domain) and recognizing (via the ankyrin repeats or chromodomain) a given methyl mark – allowing for a mechanism of crosstalk to propagate a given methyl mark. PHF8 and KIAA1718 (Fig. 5a), and perhaps JARID/Lid2 (Fig. 5b) and JMJD2A (Fig. 4b), contain modules, within the same polypeptide, for both recognizing (via the PHD or Tudor) and removing (via the Jumonji domain) two opposing methyl marks – a mechanism of crosstalk removes an “off” methyl mark based on an existing “on” methyl mark. Understanding the function and crosstalk of individual letters (one methyl mark, two methyl marks, and so on) may allow us eventually decipher the complex language of the histone code [94, 98].
The availability of human and other model research organism genome sequences, proteomics, and transcriptomics has provided answers to a wide range of questions that in some cases we did not even previously know to ask. Global analyses of genomic DNA methylation and histone modifications [99–101] are playing a similar role, yielding powerful insights into normal development and diseases, such as cancer and diabetes. The experimental characterization of individual modifying enzymes (writers) and demodifying enzymes (erasers) of the histone code is providing a growing and convergent picture of the kinetic mechanisms, binding partners, chromatin recognition, and in some cases structures of these proteins. However, it is clear that the activities of writers, erasers, and readers of the histone code are regulated in multicomponent complexes that have yet to be fully defined and characterized.
Acknowledgments
The work in the Cheng laboratory was supported by grants GM06860 and DK082678 from the National Institutes of Health (NIH). X.C. is a Georgia Research Alliance Eminent Scholar.
References
- 1.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. Embo J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 6.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 7.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 8.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 11.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 12.Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol Life Sci. 2009;66:407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 14.Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem. 2009;4:1568–1582. doi: 10.1002/cmdc.200900301. [DOI] [PubMed] [Google Scholar]
- 15.Spannhoff A, Sippl W, Jung M. Cancer treatment of the future: inhibitors of histone methyltransferases. Int J Biochem Cell Biol. 2009;41:4–11. doi: 10.1016/j.biocel.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 16.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 17.Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 18.Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J Biol Chem. 2004;279:43296–43306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian C, Zhou MM. SET domain protein lysine methyltransferases: structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couture JF, Trievel RC. Histone-modifying enzymes: encrypting an enigmatic epigenetic code. Curr Opin Struct Biol. 2006;16:753–760. doi: 10.1016/j.sbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Cheng X, Zhang X. Structural dynamics of protein lysine methylation and demethylation. Mutat Res/Fundam Mol Mech Mutagen. 2007;618:102–115. doi: 10.1016/j.mrfmmm.2006.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 25.Volkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89:1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, Selker EU, Shinkai Y, Cheng X. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couture JF, Dirk LM, Brunzelle JS, Houtz RL, Trievel RC. Structural origins for the product specificity of SET domain protein methyltransferases. Proc Natl Acad Sci USA. 2008;105:20659–20664. doi: 10.1073/pnas.0806712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi YH, Lee JS, Swanson SK, Saraf A, Florens L, Washburn MP, Trievel RC, Shilatifard A. Regulation of H3K4 trimethylation via Cps40 (Spp 1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol. 2009;29:3478–3486. doi: 10.1128/MCB.00013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Tamaru H, Khan SI, Horton JR, Keefe LJ, Selker EU, Cheng X. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111:117–127. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 32.Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Couture JF, Collazo E, Hauk G, Trievel RC. Structural basis for the methylation site specificity of SET7/9. Nat Struct Mol Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 34.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 35.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pless O, Kowenz-Leutz E, Knoblich M, Lausen J, Beyermann M, Walsh MJ, Leutz A. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-beta. J Biol Chem. 2008;283:26357–26363. doi: 10.1074/jbc.M802132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 40.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Jenuwein T. The epigenetic magic of histone lysine methylation. Febs J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- 42.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 43.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 46.Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2007;26:77–90. doi: 10.1038/sj.onc.1209763. [DOI] [PubMed] [Google Scholar]
- 47.Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, Tang C, Redkar S, Jones PA. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 2007;67:6400–6408. doi: 10.1158/0008-5472.CAN-07-0251. [DOI] [PubMed] [Google Scholar]
- 48.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, Snyder JP, Bedford MT, Cheng X. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol. 2009;16:312–317. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D. Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the jumonji domain-containing JMJD2/KDM4 proteins. J Biol Chem. 2009;284:8395–8405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 52.Xu Y, Shi Y, Ding S. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 53.Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 55.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 56.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, Kozieradzki I, Senisterra G, Chau I, Siarheyeva A, Kireev DB, Jadhav A, Herold JM, Frye SV, Arrow-smith CH, Brown PJ, Simeonov A, Vedadi M, Jin J. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a. J Med Chem. 2009;52(24):7950–7953. doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 59.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 60.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461 (7262):415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 62.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 63.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 64.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 65.Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, Ikari M, Sato M, Terada T, Nagase T, Ohara O, Shirouzu M, Tanaka A, Kigawa T, Yokoyama S. Solution structure of the SWIRM domain of human histone demethylase LSD1. Structure. 2006;14:457–468. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 66.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J Biol Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 67.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. Crystal structure of human histone lysine-specific demethylase 1 (LSD1) Proc Natl Acad Sci USA. 2006;103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 69.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 70.Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol. 2007;14:535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- 71.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J Biol Chem. 2007;282:20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 72.Trewick SC, McLaughlin PJ, Allshire RC. Methylation: lost in hydroxylation? EMBO Rep. 2005;6:315–320. doi: 10.1038/sj.embor.7400379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 74.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 75.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 76.Dann CE, 3rd, Bruick RK. Dioxygenases as O2-dependent regulators of the hypoxic response pathway. Biochem Biophys Res Commun. 2005;338:639–647. doi: 10.1016/j.bbrc.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 77.Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates JR, 3rd, Delahunty CM, Hahn P, Lengeling A, Mann M, Proudfoot NJ, Schofield CJ, Bottger A. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 78.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by the MLL fusion partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Jr, Krebs C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci USA. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 81.Tu S, Teng YC, Yuan C, Wu YT, Chan MY, Cheng AN, Lin PH, Juan LJ, Tsai MD. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat Struct Mol Biol. 2008;15:419–421. doi: 10.1038/nsmb.1400. [DOI] [PubMed] [Google Scholar]
- 82.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, Hagman J, Hansen K, Shi Y, Zhang G. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 83.Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S, Hansen K, Shi Y, Zhang G. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci USA. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 85.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, Rose NR, Offer J, Scheinost JC, Borowski T, Sundstrom M, Schofield CJ, Oppermann U. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 86.Han Z, Liu P, Gu L, Zhang Y, Li H, Chen S, Chai J. Structural basis for histone demethylation by JHDM1. Frontier Sci. 2007;1:52–67. [Google Scholar]
- 87.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 88.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ, Ratcliffe PJ, Schofield CJ. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an N {varepsilon}-dimethyl lysine demethylase. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural basis for substrate specificity of a family of Jumonji histone lysine demethylases. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 92.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 93.Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 95.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 96.Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 97.Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 98.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 99.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, Jeltsch A. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet. 2009;5:e1000438. doi: 10.1371/journal.pgen.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 103.Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 2005;19:1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, Wilson JR. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 2005;19:1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]