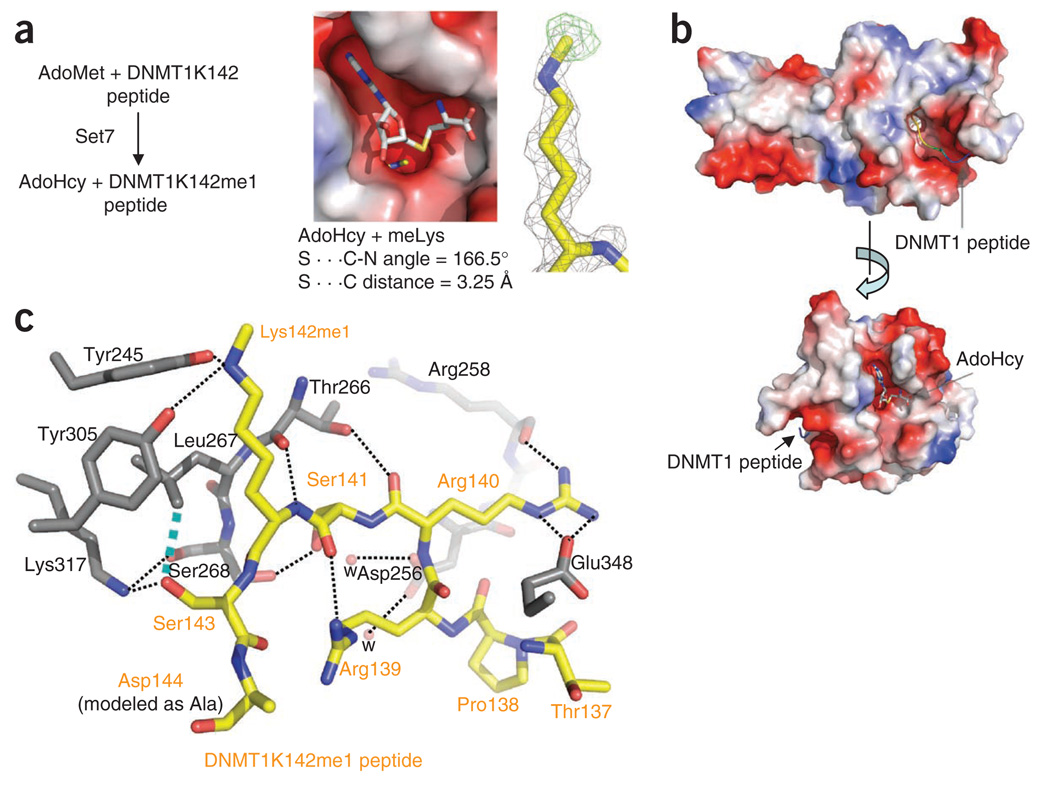
Figure 2.
Structure of the SET7–DNMT1 complex. (a) The reaction occurred during crystallization. Omit electron densities, Fo − Fc contoured at 4σ above the mean, are shown for the monomethylated Lys142 (black mesh) and the methyl group (green mesh). (b) The substrate peptide and the reaction product AdoHcy occupy binding sites in the opposite ends of a narrow target-lysine channel. (c) Electrostatic interactions, hydrogen bonds and van der Waals interactions define SET7–DNMT1 peptide interactions (dashed lines). The network of interactions includes the following: (i) DNMT1 Arg139 forms a hydrogen bond with the main chain carbonyl oxygen atom of SET7 Gly336 (not shown) and an intramolecular interaction with the main chain carbonyl oxygen of DNMT1 Ser141; (ii) DNMT1 Arg140 forms an electrostatic salt bridge to SET7 Glu348 and a hydrogen bond with the main chain carbonyl oxygen atom of SET7 Arg258; (iii) DNMT1 Ser141 forms a serineserine interaction with SET7 Ser268 and a water-mediated network with DNMT1 His252 (not shown) and SET7 Asp256; (iv) the target nitrogen atom of DNMT1 Lys142me1 forms two hydrogen bonds with the side chain hydroxyl oxygen atoms of SET7 Tyr305 and Tyr245; and (v) DNMT1 Ser143 is involved in a polar interaction with SET7 Lys317 and a van der Waals contact with SET7 Leu267. In addition, two main chain atoms of DNMT1, the carbonyl oxygen of Arg140 and the amide nitrogen of Lys142me1, link through SET7 Thr266.