Abstract
Mek1 is a Chk2/Rad53/Cds1-related protein kinase that is required for proper meiotic progression of Schizosaccharomyces pombe. However, the molecular mechanisms of Mek1 regulation and Mek1 phosphorylation targets are unclear. Here, we report that Mek1 is phosphorylated at serine-12 (S12), S14 and threonine-15 (T15) by Rad3 (ATR) and/or Tel1 (ATM) kinases that are activated by meiotic programmed double-strand breaks (DSBs). Mutations of these sites by alanine replacement caused abnormal meiotic progression and recombination rates. Phosphorylation of these sites triggers autophosphorylation of Mek1; indeed, alanine replacement mutations of Mek1-T318 and -T322 residues in the activation loop of Mek1 reduced Mek1 kinase activity and meiotic recombination rates. Substrates of Mek1 include Mus81-T275, Rdh54-T6 and Rdh54-T673. Mus81-T275 is known to regulate the Mus81 function in DNA cleavage, whereas Rdh54-T6A/T673A mutant cells showed abnormal meiotic recombination. Taken together, we conclude that the phosphorylation of Mek1 by Rad3 or Tel1, Mek1 autophosphorylation and Mus81 or Rdh54 phosphorylation by Mek1 regulate meiotic progression in S. pombe.
Key words: Mek1, meiotic recombination, phosphorylation, Rdh54, Mus81
Introduction
In many organisms that reproduce sexually, meiosis entails a single round of DNA replication followed by homologous recombination (HR) and two successive rounds of chromosome segregation, a process that increases the genetic diversity of the offspring and promotes the survival of the species during critical environmental changes.1,2 HR plays an essential role in repairing DSBs that arise unpredictably by damaging agents or that are induced in a programmed manner during meiosis.3 Unlike mitotic recombination, DSB repair during meiotic recombination involves homologous chromosomes, as well as sister chromatids, as repair templates. In budding yeast Saccharomyces cerevisiae this partner choice is partly created by the meiosis-specific paralog of the Rad53 effector kinase, Mek1.4 Rad53 is required for the DNA damage checkpoint response to accidental DSBs that occur during mitotic devisions, whereas Mek1 (together with Red1 and Hop1) promotes recombination between homologous chromosomes by suppressing DSB repair between sister chromatids during meiosis. Rad53 cannot transduce meiosis-specific DSB signals, probably due to its failure to access meiotic recombination sites.5 Meiosis-induced opening of chromatin is associated with meiotic DSB hotpots6 and chromosome axes are structurally modified at future crossover sites.7 Moreover, the Red1-SUMO chain interaction is essential for Tel1- and Mec1-dependent Hop1 phosphorylation, which ensures interhomolog recombination (IHR) by preventing the inter-sister chromatid DNA repair pathway.8
Mek1 kinase activity of S. cerevisiae is required for normal meiosis, as shown by the aberrant exit of the mek1 kinase-dead mutant from the pachytene stage of meiotic prophase and by its reduced spore viability, equivalent to that of the mek1 null mutant.9 Mek1 forms a complex with the meiosis-specific chromosomal components Red1 and Hop1, which are required for full kinase activity of the Mek1.10 Mek1 is activated in response to DSBs by autophosphorylation of two conserved threonine residues, T327 and T331, in the Mek1 activation loop, and this phosphorylation is necessary for the maintenance of Mek1 dimers during checkpoint-induced arrest.11 Mec1 and Tel1, the budding yeast homologs of the mammalian ATR and ATM kinases, also promote meiotic HR by phosphorylation of the axial element protein Hop1, and this modification is essential for Mek1 activation.12
In Schizosaccharomyces pombe, homologous proteins are also required for meiotic recombination and the meiotic recombination checkpoint.2,13 Intrachromosomal and sister chromatid recombination are important, especially in S. pombe, and may be more frequent than recombination between homologs.14,15 Cds1 (Rad53) plays an important role in the mitotic cell cycle as the major effector of the replication checkpoint.16 Cds1 and its orthologs in other species (S. cerevisiae Rad53 and mammalian CHK2) are characterized by an N-terminal Ser-Gln/Thr-Gln (SQ/TQ) cluster and forkhead-associated (FHA) domain and a Ser-Thr kinase domain.17,18 Cds1 activation requires the upstream kinase Rad3 (Mec1/ATR) and the mediator Mrc1. An FHA domain interaction with Mrc1 mediates Cds1-T11 phosphorylation by Rad3; this phosphorylation is critical for Cds1 function.19,20 Activation occurs as follows: first, Mrc1 recruits Cds1 to stalled replication forks by interactions between the Cds1 FHA domain and specific phosphorylated Rad3 consensus sites in Mrc1, and then Cds1 dimerizes via phospho-specific interactions mediated by the FHA domains and is activated by autophosphorylation.21 Rad3-Cds1 pathway coordinates the initiation of meiotic recombination and meiotic cell divisions with premeiotic DNA synthesis.22 Mek1 plays a pivotal role in the meiotic recombination checkpoint23,24 and appears to enhance homolog interactions to assure WT levels of crossing over.15 However, knowledge about the position and role of S. pombe Mek1 in the signal transduction cascades during meiosis is limited.
The FHA domain of Cds1 interacts with the heterodimeric endonuclease Mus81-Eme1, a Holliday junction resolvase,15 which is required for meiotic crossovers in S. pombe.25–27 Rdh54 (Tid1), a member of the Swi2/Snf2 family of chromatin remodeling proteins, stimulates Dmc1-dependent meiotic recombination and promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin.28 Processive and ATP-dependent translocation of Rdh54 on DNA displaces associated proteins and migrates recombination intermediates.29 Rdh54 is expressed during meiosis and acts with Rhp54 (a Rad54 ortholog) to repair meiotic DSBs through interactions involving Rhp51 and the meiotic Rhp51 homolog Dmc1.30,31 However, little is known about the interaction of Rhp54 with Mek1.
Here, we report studies on the regulation and function of Mek1 during meiosis. We report that the Rad3 and/or Tel1 phosphorylate Mek1 on the S12, S14 and T15 residues, and that these modifications are required for meiotic progression. In particular, phosphorylation of Mek1-T15 is important for the initiation of meiotic S phase. Mek1 then activates itself by autophosphorylation at Mek1-T318. Subsequently, activated Mek1 phosphorylates Mus81 on T275, which negatively regulates the Mus81 function.32 Furthermore, Mek1 phosphorylates Rdh54 on T6 and T673, which appears to have a regulatory role in meiotic progression.
Results
Rad3 and Tel1 phosphorylate Mek1 S12, S14 and T15 in vitro.
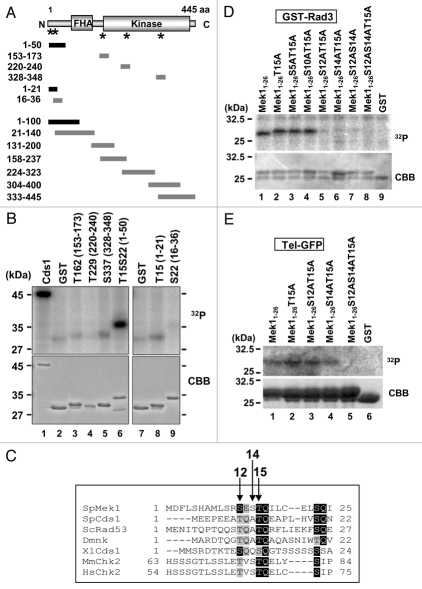
We previously showed that Mek1 phosphorylation was induced during early meiosis to initiate meiotic recombination.23 Since activation of Cds1 following DSB formation depends on its phosphorylation by Rad3, we surmised that Rad3 also phosphorylates Mek1. Indeed, Mek1 has five SQ/TQ motifs that are preferentially phosphorylated by PIK-like kinases such as Rad3 (Fig. 1A and asterisks). Thus, we examined whether any of these motifs serves as a Rad3 phosphorylation target by performing in vitro kinase assays using affinity-purified GST-fused Mek1 fragments with specific SQ/TQ motifs as substrates (T162, T229, S337 and T15S22). GST-Cds1 and GST alone were used as positive and negative controls, respectively. Among the four substrates tested, the GST-fused peptide fragment containing T15S22 most strongly incorporated 32P (Fig. 1B and lane 6). To determine whether T15 or S22 is phosphorylated, the T15S22 region was subcloned and the T15 and S22 peptide fragments were subjected to the kinase assay, which showed that the T15 fragment incorporated 32P more strongly than did the S22 fragment (Fig. 1B and lane 8). Moreover, addition of wortmannin, an inhibitor of ATM/ATR and Rad3 kinase activity, reduced the amount of 32P incorporation in a dose-dependent manner (Sup. Fig. 1A). To determine if Mek1 Ser or Thr residues other than those in SQ/TQ motifs are phosphorylated by Rad3, seven plasmids that express GST-Mek1 fragments that cover the whole Mek1 molecule were constructed (Fig. 1A). In vitro kinase assays showed that Rad3 phosphorylated only the GST-Mek1 fragment containing amino acids 1–50 (Sup. Fig. 1B). Thus, the phosphorylation sites are located between amino acids 1–21 of Mek1.
Figure 1.
Rad3 and Tel1 phosphorylate S12, S14 and T15 in the N-terminal region of S. pombe Mek1. (A) Structure of S. pombe Mek1 (455 amino acids) with FHA and kinase domains. Asterisks indicate the locations of motifs that are commonly found around the S and T phosphorylation targets. (B, D and E) GST-Rad3 in (B and D) or Tel-GFP in (E), phosphorylates GST-Mek1 fragments, as revealed by in vitro kinase assays. Proteins were separated by SDS-PAGE, and the kinase assay substrates are indicated above each lane. Incorporation of 32P is shown in the upper part (32P), and the amount of protein assessed by Coomassie brilliant blue (CBB) staining is shown in the lower part. 32P incorporation was visualized using a phosphor image analyzer (Fuji Film, Tokyo, Japan). (C) Alignment of the N-terminal SQ/TQ cluster domains of the fission yeast Mek1 (SpMek1) and Cds1 (SpCds1), budding yeast Rad53 (ScRad53), Drosophila melanogaster Dmnk, Xenopus laevis Cds1 (XlCds1), Mus musculus Chk2 (MmChk2) and Homo sapiens Chk2 (HsChk2). Hyphens represent gaps that were inserted to maximize homology. Amino acids identical to those in Mek1 are shaded in black, while similar residues are shaded in gray.
The alignment of amino acid sequences of members of the CHK2 protein kinase family, which includes Mek1, showed that T15 is conserved in all proteins examined except for S. cerevisiae Mek1 and C. elegans Cds1 (Figs. 1C and Sup. Fig. 1C). Since S5, S10, S12, and/or S14 of Mek1 could also be phosphorylation targets, we constructed plasmids that express Mek1 mutants in which these S/T residues are replaced by alanine (A). In vitro kinase assays with Rad3 showed that phosphorylation was reduced by the S12A, S14A or T15A mutations, and the peptide fragment harboring S12A/S14A/T15A incorporated negligible amounts of 32P (Fig. 1D and lane 8). These results indicate that Rad3 phosphorylates these Mek1 residues, at least in vitro. Similarly, the Tel1 kinase, a Rad3 homolog, also phosphorylates these residues in vitro (Fig. 1E and lane 5).
Phosphorylation of Mek1 in vivo during meiosis.
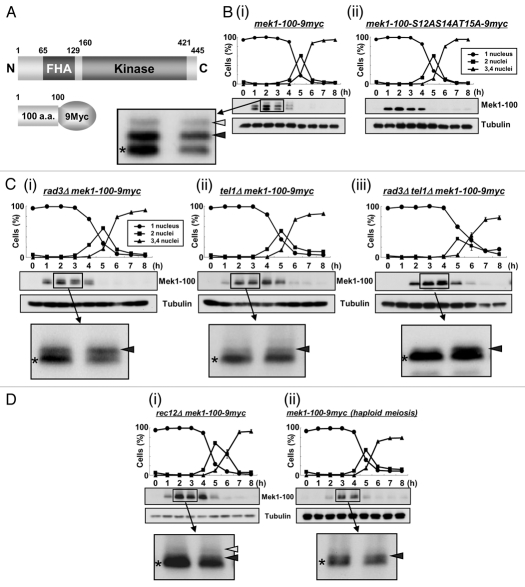
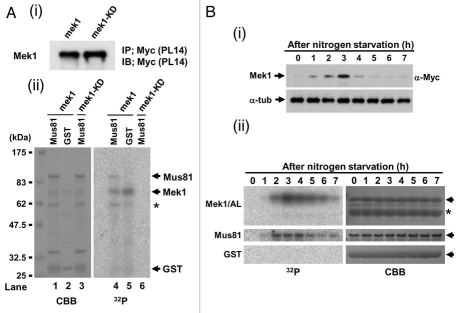
We next examined Mek1 phosphorylation profiles in vivo by western blot analysis. Since the extent of band shift detected by SDS-PAGE was too small to be distinguished by non-shifted band, we constructed an S. pombe strain that expresses a 9Myc-tagged Mek1 fragment (amino acids 1–100) from its native promoter (TT623) (Fig. 2A), which allowed visualization of two shifted bands during meiosis (Fig. 2Bi). An S. pombe mutant strain that expresses a 9Myc-tagged Mek1-100-S12AS14AT15A mutant protein (TT710) during meiosis served as a negative control. Indeed, the shifted bands (white and black arrowheads in Fig. 2Bi) disappeared in the Mek1-S12A/S14A/T15A mutant cells (Fig. 2Bii), which indicates that they are due to phosphorylation of Mek1 S12, S14 or T15, and that the lower band (asterisk) that was observed in both WT and mutant cells is the unphosphorylated Mek1 fragment. Examination of these bands in the rad3Δ or tel1Δ genetic backgrounds (TT705/TT286/TT201) revealed that the uppermost band (white arrowhead) almost disappeared in these mutant cells (Fig. 2C), which suggest that this band is derived from phosphorylation by Rad3 or Tel1. In contrast, the intensities of the lower band (black arrowhead) remained almost unchanged. The intensity of upper band was reduced in tel1Δ (TT286) cells compared to rad3Δ (TT705) cells (Fig. 2Ci and Cii), suggesting a greater role for Tel1 in Mek1 phosphorylation. Notably, although most of the band shifts disappeared at 2–3 h after meiotic induction in rad3Δtel1Δ cells (TT201), a new band shift was detected at 4–6 h, suggesting that an unknown kinase participated in Mek1 phosphorylation when both Rad3 and Tel1 were disrupted (Fig. 2Ciii).
Figure 2.
Mek1 is phosphorylated in vivo during meiosis. (A) Schematic of the truncated Mek1-100-9Myc protein fusion. Plasmids carrying the Mek1-100-9Myc fusion construct or a construct with the S12AS14AT15A mutations were integrated into the mek1 promoter locus in mek1Δ cells. (B) Detection of Mek1 bands in mek1-100-myc cells (TT623) (i) or mek1-100-S12AS14AT15A-9myc cells (TT710) (ii) during meiosis by western blot analysis. Only a single band is detected in TT710 cells, in which S12, S14 and T15 of Mek1 are replaced by alanine. (C) Detection of multiple bands of Mek1 in rad3Δ mek1-100-9myc cells (TT705) (i), tel1Δ mek1-100-9myc cells (TT286) (ii) or rad3Δ tel1Δ mek1-100-9myc cells (TT201) (iii) during meiosis. Asterisks indicate putative unphosphorylated bands, while white and black arrowheads denote putative phosphorylated bands. The intensity of the black arrowhead band is remarkably weakened in tel1Δ mek1-100-9myc cells, suggesting it to be primarily derived from phosphorylation by Tel1. (D) Detection of multiple bands of Mek1 during meiosis in rec12Δ mek1-100-9myc cells (TT266) (i) or in haploid mek1-100-9myc cells (TT608) (ii). The rec12Δ cells cannot generate double strand breaks during meiosis, while the mek1-100-9myc cells were induced to undergo haploid meiosis in the pat1 genetic background. (E) Hydroxyurea (HU) blocks DNA replication of mek1-100-9myc cells, as observed by the persistence of single-nucleus cells and by FACS analysis. (F) Detection of multiple Mek1 bands in mek1-100-9myc cells after HU treatment during meiosis.
DSB formation is required for the initiation of meiotic recombination, and S. pombe rec12+, which encodes a protein homologous to S. cerevisiae Spo11, is essential for DSB generation.33–36 In rec12Δ cells (TT266), which cannot produce DSBs during meiosis, the intensity of the uppermost band of Mek1 (white arrowhead) was reduced (Fig. 2Di) compared with WT cells (Fig. 2Bi), which indicates a tight link between Mek1 phosphorylation and DSB formation. A slower rate of DSB formation and a reduction in the frequency of DSBs are observed in haploid meiosis, in which a homologous partner for recombination is absent. Indeed, the intensities of the shifted band (white arrowhead) was remarkably reduced during haploid meiosis in mek1-100-9myc cells (TT608) (Fig. 2Dii), which also supports the tight linkage of these phosphorylated bands to DSB formation. Nonetheless, Mek1 phosphorylation is not required for recombination itself (see Sup. Fig. 5D).
Notably, Mek1 expression was delayed until 4 h after meiotic induction in cells lacking Rad3 or Tel1 (TT551/TT552/TT553) when the actions of Rad3 and Tel1 with respect to Mek1 and meiotic recombination in the horsetail phase were almost complete in mek1+-9myc cells (TT305) (Sup. Fig. 2A). Specifically, Mek1 was not expressed in a timely manner in the absence of Rad3 and Tel1, suggesting that the timing of Mek1 expression is partly regulated by Rad3 and Tel1, and that Mek1 is expressed at 4 h to carry out a Rad3/Tel1-independent function. Moreover, Mek1 expression was slightly enhanced at 4 and 5 h, even in rad3Δtel1Δ cells (TT553), which further supports the idea that an unknown kinase also phosphorylates and regulates Mek1 (Fig. 2Ciii).
Mek1 kinase activity in these genetic backgrounds was investigated by immunoprecipitating Mek1-9Myc 4 h after meiotic induction and performing an in vitro kinase assay using the activation loop region of Mek1 as a substrate (see Fig. 4B). The kinase activity of Mek1 was remarkably reduced in rad3Δtel1Δ cell extracts (TT553). These data suggest that Mek1 kinase activity is upregulated by Rad3 or Tel1 phosphorylation (Sup. Fig. 2B).
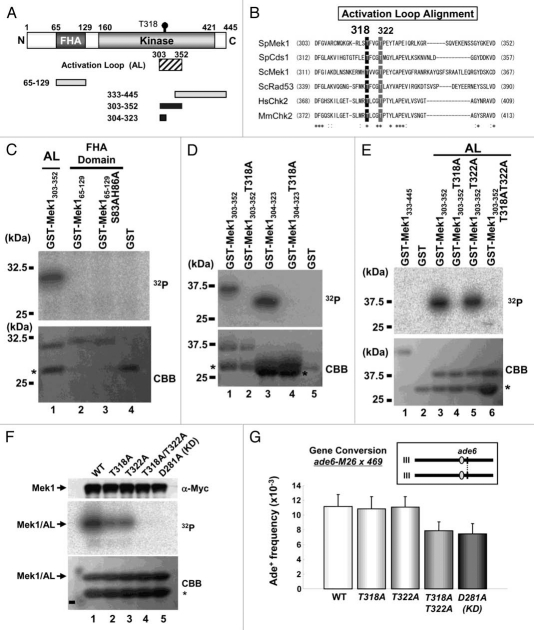
Figure 4.
Mek1-T318 and -T322 are necessary for the full kinase activity of Mek1 and meiotic recombination. (A) Schematic diagram of S. pombe Mek1 shows the locations of the Activation Loop (AL; shaded box) and four fragments (black and gray boxes) used as substrates for the in vitro kinase assay. Black and gray colors indicate phosphorylated and non-phosphorylated fragments, respectively. Mek1 fragment amino acid numbers are shown at the left of each box. (B) Alignment of the primary structure around the conserved T318 and T322 residues (highlighted) of S. pombe (Sp) Mek1 with other related protein fragments. Asterisks or colons signify identical or almost identical amino acids among the denoted proteins. (C–E) Mek1 kinase (Mek1-9Myc immunoprecipitate) phosphorylates the Mek1 fragments indicated above each lane. Upper parts, incorporation of 32P; lower parts, the amount of loaded protein as assessed by CBB staining. Replacements of the S83, H86, T318 and T322 residues by alanine (A) are denoted. Asterisks indicate putative degradation products. (F) Detection of the kinase activity of Mek1 mutants in vitro. WT and Mek1 mutant proteins were prepared by immunoprecipitation, and their kinase activities were measured using the AL fragment as a substrate. The amount of immunoprecipitated Mek1 kinases assessed by western analysis with the anti-Myc antibody, the incorporation of 32P into the tested substrate, and the amount of loaded protein assessed by CBB staining are shown in the upper, middle and lower parts, respectively. The lower bands in the lower part are degradation products (asterisk). (G) Intragenic recombination between the ade6-M26 and ade6-469 alleles on chromosome III in WT (TT230 × TT231), mek1-T318A (TT677 × TT678), mek1-T322A (TT679 × TT680), mek1-T318AT322A (TT681 × TT682) or mek1-D218A (TT591 × TT592) strains. The positions of the ade6+ locus and centromeres (gray balls) on chromosome III are illustrated in the inset. The data show the averages and standard deviation of triplicate experiments.
Meiotic phenotypes of Mek1 mutants.
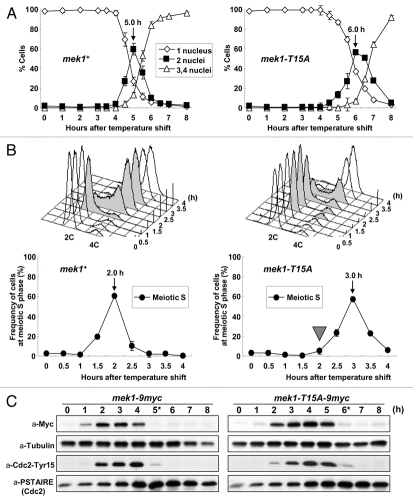
Of the Mek1 phosphorylation sites, Mek1-T15 appears to be most important because it corresponds to the previously reported phosphorylation site of Cds1-T11, while S12 and S14 are not conserved in Cds1. Thus, to identify the physiological consequence of Mek1-T15 phosphorylation, we first examined meiotic progression in the mutant strain mek1-T15A by time course FACS analysis and detected a substantial (1 h) delay in the initiation of meiotic S phase (Fig. 3A and B), which was reflected by the prolonged phosphorylation of Tyr15 of Cdc2 (Fig. 3C). This phenotype is noteworthy considering that mek1Δ cells showed no delay in the initiation of meiotic S phase.15
Figure 3.
Meiotic progression is delayed in mek1-T15A-9myc pat1 cells. (A) Meiotic progression was monitored by counting nuclei following the induction of meiosis at the indicated times in mek1+ cells, mek1-9myc pat1 cells (TT305) and mek1-T15A-9myc pat1 cells (TT273). The data are the means ± SD of at least three independent experiments. (B) Progression of meiotic S phase was monitored by FACS analysis of the DNA content of samples collected at each time point (upper parts). The fraction of cells undergoing DNA replication at each time point was determined from the FACS profiles with Modfit LT software (lower parts). (C) Western blot detection of Mek1-9myc, α-Tubulin, Tyr15 phosphorylated Cdc2 and total Cdc2. Samples were taken after the temperature shift at the indicated times. α-tubulin was used as a loading control. Asterisks show the time at which the number of cells possessing two nuclei peaked.
Next, we analyzed four mek1 mutants, mek1-KD (GP21), -FHAD (TT277), -T15A/KD (TT526) and -T15A/FHAD (TT548); the KD mutant lacks kinase activity and the FHAD mutant lacks the FHA domain function. Notably, the mek1-KD strain (GP21) was not delayed in meiosis (Sup. Fig. 3), while the mek1-FHAD strain displayed a 30 min delay, which is intermediate between those of the mek1-T15A (TT273) and mek1-KD mutants (Sup. Fig. 4). Further, mek1-T15A/KD and -T15A/FHAD mutants also showed 30 min delays (Figs. S3 and S4). These results indicate that Mek1-T15 phosphorylation is important for the initiation of meiotic S phase. It also suggests that Mek1-T15 phosphorylation plays a role in the regulation of meiotic recombination checkpoint.23
The effect of mek1 mutations on HR was investigated by determining the rates of intergenic and intragenic recombination in mek1-KD, mek1-FHAD and mek1-T15A cells (TT188 × TT189, TT164 × TT165 and TT162 × TT163), as well as in double mutant cells (Sup. Fig. 5A and B). The recombination frequencies of mek1Δ cells (TT253 × TT254) were substantially reduced (40% and 36% of WT), while those of mek1-KD cells (TT188 × TT189) were reduced to a lesser extent (56% and 59% of WT). In contrast, the recombination frequencies of mek1-T15A cells (TT162 × TT163) were almost normal, while those of the mek1-FHAD mutant (79% and 64% of WT) were intermediate between those of the mek1-T15A and mek1Δ mutants. The presence of the T15A mutation in mek1-KD and mek1-FHAD cells (TT475 × TT476 and TT541 × TT542) had little effect on meiotic recombination frequencies. This result indicates that the Mek1 kinase activity plays a role in meiotic recombination. In contrast, the FHA domain is partially involved, but the T15A mutation has little effect on meiotic recombination.
The potential involvement of the S12 and/or S14 mutations in recombination was also examined, and it was found that mek1-S12A, mek1-S14A and mek1-T15A cells (TT689 × TT690, TT691 × TT692, TT162 × TT163) had normal meiotic recombination rates (Sup. Fig. 5B; see also 5D). Moreover, even the triple alanine (mek1-3A) mutant (TT675 × TT676), in which S12, S14 and T15 were replaced by alanine, showed only a slight decrease in the gene conversion rate compared to that in WT cells (TT230 × TT231) (88% of the WT level). Inclusion of the mek1-3A mutation in mek1-FHAD cells also had little effect on the already decreased gene conversion rate. These results indicate that phosphorylation of the S12, S14 and T15 residues of Mek1 is irrelevant to its meiotic recombination function. Finally, the spore viability of these mutant cells was not remarkably different from that of WT cells (TT230 × TT231) (Sup. Fig. 5C).
Mek1-T318 and -T322 are required for Mek1 activation through autophosphorylation.
Human Chk2 and Cds1 are activated by autophosphorylation.21,37 Since the primary structures of the FHA domain and activation loop (AL) region (Fig. 4A) are highly similar among Mek1 orthologs—in particular, the putative autophosphorylation targets are conserved (Fig. 4B)—it is likely that Mek1 is also activated by phosphorylation of the corresponding residues. This hypothesis was tested by preparing GST-fused Mek1-AL and Mek1-FHA fragments including amino acids 303–352 and 65–129, respectively, as substrates. These were examined in an in vitro kinase assay, which showed that only the Mek1-AL fragment underwent autophosphorylation (Fig. 4C). Two Mek1-T318A mutant proteins, Mek1303–352 and Mek1304–323, were next prepared and tested, and both were found to be unphosphorylated (Fig. 4D). This result suggests that Mek1-T318 is the autophosphorylation target in vitro. To confirm that the rest of the C-terminal domain of Mek1 including the Mek1-T322 residue do not undergo phosphorylation, we also prepared and tested a Mek1333–445 fragment and a Mek1303–352 fragment harboring the Mek1-T322A mutation (Fig. 4E). The former was not phosphorylated and the latter exhibited phosphorylation signals (Fig. 4E and lanes 1 and 5), which supports the conclusion that the T318 is the major autophosphorylation target in vitro.
Next, the T318A and/or T322A point mutations were examined to determine if they affect the kinase activity of Mek1 in vivo. S. pombe strains that express the 9Myc-tagged Mek1 (WT), Mek1-T318A, Mek1-T322A, Mek1-T318AT322A and Mek1-D281A (kinase-dead; KD) proteins under the control of the nmt41 promoter were constructed. Extracts of each cell type in the vegetative growth phase were prepared and immunoprecipitated with the anti-Myc antibody, and kinase activities of the precipitates were measured using the GST-fused AL fragment as a substrate. As shown in Figure 4F, the T318A or T322A single mutation reduced Mek1 kinase activity to similar levels, while the T318A/T322A double mutation abolished Mek1 kinase activity, as did the kinase-deficient (D281A) mutation. These results indicate that both T318 and T322 are necessary for full Mek1 kinase activity in vivo, as seen for other CHK2 family kinases.37 Namely, this suggests that Mek1-T322 may participate in activation of Mek1 after autophosphorylation occurred at Mek1-T318.
We next examined the meiotic recombination rates in mek1-9myc (WT), mek1-T318A-9myc, mek1-T322A-9myc, mek1-T318AT322A-9myc and mek1-D281A-9myc (kinase-dead mutant) cells (TT230 × TT231, TT677 × TT678, TT679 × TT680, TT681 × TT682 and TT591 × TT592). The mek1-T318A-9myc and mek1-T322A-9myc mutants were unchanged compared with mek1-9myc cells. In contrast, the gene conversion rate was modestly reduced in mek1-T318AT322A-9myc mutant cells (70% of WT), to the level of the mek1-D281A-9myc mutant (Fig. 4G). These results support the conclusion that both T318 and T322 are necessary for the full kinase activity of Mek1 in vivo and play a role in meiotic recombination.
Mek1 associates with and directly phosphorylates Mus81.
Since Cds1 specifically interacts with and phosphorylates Mus81,25,34 we examined if Mek1 also phosphorylates Mus81 in vitro. To avoid contamination of unknown meiotic protein kinase, Mek1 kinase activity was examined by preparing immunoprecipitates of cell extracts from logarithmically growing (mitotic) S. pombe cells that expressed Mek1-9Myc or Mek1-KD-9Myc proteins from the nmt41 promoter (pREP41; see Sup. Fig. 2), and their equal expression levels were confirmed by western blot analysis using the anti-Myc antibody (Fig. 5Ai). The Mek1-9Myc immunoprecipitate (Fig. 5Aii): lane 4) phosphorylated GST-Mus81 but not GST alone (Fig. 5Aii): lane 5). In contrast, the Mek1-KD-9Myc immunoprecipitate failed to phosphorylate GST-Mus81 (Fig. 5Aii): lane 6). Immunoprecipitates from meiotic S. pombe cells were next examined for their ability to phosphorylate GST-Mus81, with respect to the Mek1 expression level, which peaks after 3 h of nitrogen starvation (Fig. 5Bi). Indeed, meiotic Mek1-9Myc immunoprecipitates also phosphorylated GST-Mus81 (but not GST alone) with a peak around 2–4 h, a timing similar to that of the phosphorylation of the Mek1/AL fragment, which was used as a positive control (Fig. 5Bii): left parts). These results suggest that Mek1 directly phosphorylates Mus81 at least in vitro.
Figure 5.
Mek1 and/or Cds1 phosphorylate the T218, T275 and/or T422 residues of S. pombe Mus81 in vitro. (A) Schematic diagram of GST-Mus81 fragments. Mus81 fragment amino acid numbers are shown left of each box. (B ∼ E) refers to fragments whose in vitro kinase assay data are shown below in (B ∼ E). Black and gray boxes indicate phosphorylated and non-phosphorylated fragments, respectively. Sub1 (substrate 1) and Sub2 (substrate 2) signify the GST-Mus81 fragments that were used for the kinase assay in (B). (B–F) SDS-PAGE parts for in vitro kinase reactions performed with truncated GST-Mus81 fragments and Mek1-GFP (i) or Cds1-2HA (ii) as kinases. Simply Blue staining (SB, left parts, C–F) shows a loading control. (B) The truncated GST-fusion proteins used were those containing Sub1 and Sub2, and derivatives with replacement of the T218 and T422 residues by alanine (A). (C) The truncated GST-fusion proteins used were those containing amino acids 1–127, 32–217, 219–421 and 423–608 of Mus81. An asterisk shows the autophosphorylated band of Mek1 kinase. (D) The truncated GST-fusion proteins used were those containing amino acids 219–421, 219–281, 252–362 and 336–421 of Mus81. (E) The truncated GST-fusion proteins used were those containing amino acids 252–362, 252–313, 287–362 and 317–362 of Mus81. (F) The truncated GST-fusion proteins used were those containing amino acids 252–281 and 276–313 of Mus81, and derivatives with replacement of T260, S270 and T275 residues by (A). GST-252-281 (3A) signifies that this GST-Mus81 fragment harbors all of these replacements.
Next, we determined that Mek1 and Cds1 phosphorylate Mus81 at T218, T275 and T422 in vitro (Sup. Fig. 6; see Suppl. Results for details). The reduced electrophoretic mobility of Mus81 due to T275 phosphorylation was retained in mek1Δ cells (TT464), although it was completely abolished in cds1Δ cells (TT468) (Sup. Fig. 8). Thus, the contribution of Mek1-induced Mus81-T275 phosphorylation to meiosis may be small, if there is any.
Mek1 appears to be primarily responsible for the T218 and T422 phosphorylations of Mus81 as judged by the greater efficiency of 32P incorporation by Mek1 than by Cds1, at least in vitro, when compared with the relative intensities of the Simply Blue-stained bands (white arrows and arrowheads in Sup. Fig. 7C–F) and the phosphorylated bands (black arrows and arrowheads in Sup. Fig. 7C–F). To determine the physiological significance of these phosphorylations on meiosis, we prepared mus81.T218A. T422A double mutants cells (denoted as TATA in Sup. Fig. 8) and examined their meiotic recombination rates and spore viability. Unfortunately, we could not properly compare the gene conversion rates because that of mus81-9myc was already reduced to the level of mek1-KD cells (TT591 × TT592) (Sup. Fig. 8A). The mus81.T218A.T275A.T422A triple mutant cells (denoted as 3A in Sup. Fig. 8) were also similar to mus81-9myc cells in this respect, whereas the rate of crossing over was unaltered among these mutants (Sup. Fig. 8B). Spore viabilities were also unaltered among these mutant cells (Sup. Fig. 8C). We prepared an anti-Mus81-pT218 antibody that recognized only the phosphopeptide; however, no epitope specific signal was obtained by western blotting and immunocytochemistry during S. pombe meiosis (data not shown). Thus, the physiological significance of phosphorylations on Mus81-T218 and -T422 remains unclear.
Mek1 phosphorylates Rdh54-T6 and T673.
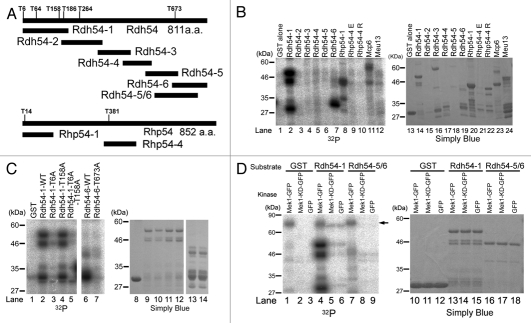
We previously reported that the HR frequencies of the mek1+ deletion mutant and the Mek1 kinase-dead mutant (Mek1-KD) are largely reduced as compared with that of WT cells,23 which suggests that Mek1 phosphorylates recombination factors and promotes HR. To identify the putative phosphorylation target(s) of Mek1, we screened recombination proteins for the Mek1 consensus target motif (RXXT). Four candidates (Rdh54, Rhp54, Mcp6 and Meu13) were selected, and in vitro Mek1 kinase assays were performed using the corresponding GST fusions of these fragments as substrates. Since preliminary data showed that the full-length Rdh54 protein is unstable (data not shown), Rdh54 was subdivided into six fragments and GST fusion proteins were prepared with these fragments (Fig. 6A). For in vitro kinase assays, extracts were prepared from mitotic S. pombe cells that harbor plasmids expressing Mek1-GFP from the nmt1 promoter. These extracts were immunoprecipitated with the anti-GFP antibody, and the precipitates were used to test the Mek1 kinase candidates. Mek1 phosphorylated the N-terminus of Rdh54 (Rdh54-1), the C-terminus of Rdh54 (Rdh54-6), and the N-termini of Rhp54 (Rhp54-1) and Mcp6/Hrs1,38,39 but it did not phosphorylate Meu13 (Fig. 6B).40
Figure 6.
Mek1 phosphorylates Rdh54 on T6 and T673. (A) Schematic presentation of the RXXT site and the fragment map of Rdh54 and Rhp54 derivatives that are expressed as N-terminal GST-fused proteins. (B) Left part, Mek1 kinase assay performed in vitro with Mek1-GFP immunoprecipitates from mitotic S. pombe cells and the GST-fused Rdh54 and Rhp54 fragments, and Mcp6 and Meu13 proteins as substrates. Right part, Simply Blue staining of an SDS-PA gel shows a loading control. (C) Mek1 kinase assay performed in vitro with GST-fusion Rdh54 truncated proteins with/without point mutations, in which T6, T158 and/or T673 were replaced with alanine. (D) Mek1 kinase assay performed in vitro with immunoprecipitates of Mek1-GFP (positive control), Mek1-KD-GFP (kinase-dead form) or GFP (negative control), and truncated GST-fused Rdh54-1 or Rdh54-5/6 fragments or GST alone as substrates. The arrow indicates autophosphorylation of Mek1. (E) Western blot analysis conducted during S. pombe meiosis using an anti-Rdh54-pT6 antibody in the absence (i) or presence (ii) of a phosphopeptide (KRRApTFQCPLIEC) that was used as an antigen. The h−/h− pat1-114 mug28+-3HA strain was induced to enter meiosis synchronously by a temperature shift, and cells were collected at 1 h intervals for protein extraction, blotting and probing with the anti-Rdh54-pT6 antibody. Tilted arrowheads in (i) indicate putative Rdh54-pT6 bands that disappeared after a competition experiment, in which the phosphopeptide was incubated with the antibody before probing the western blot, as indicated by tilted arrows in (ii). Asterisks indicate putative nonspecific bands that were not weakened after peptide competition. Meiotic expression of Meu13 was analyzed to identify the meiotic stage at each time point. Cdc2 and α-tubulin levels were examined as loading controls. (F) Dot blot analysis indicates that the anti-Rdh54-pT6 antibody recognized the phosphopeptide that was used as an antigen, but not the non-phosphopeptide (KRRATFQCPLIEC). (G) Gene conversion rates involving ade6-M26 and ade6-469 on chromosome III were measured in rdh54 point mutants. The bar graph with error bars shows the average ± SD values of three independent experiments. (H) Observation of Rdh54 nuclear foci in rdh54 mutant cells and the mek1Δ strain. The rdh54-WT (TK118-WTG), rdh54-T6A (TK118-T6A-1), rdh54-T6A/T673A (TK201-1), rdh54-K241R (TK119-K241R-1), rdh54-T6A/K241R (TK118-T6A/K241R-1) and mek1Δ (TK123) cells induced to enter meiosis were analyzed by fluorescence microscopy for Rdh54-GFP (green). Typical images are shown in the upper parts. Scale bar: 10 µm. The bar graph shows the frequency of cells with 0, 1–3, 4–7 or 8–12 Rdh54-GFP foci at the horsetail phase.
Since Rdh54-1 and Rdh54-6 carry four and one RXXT motifs, respectively (Fig. 4A), these fragments were subjected to further analysis to determine which RXXT motifs are actually phosphorylated by Mek1. The T6 and T158 residues of Rdh54-1 and the T673 residue of Rdh54-6 were replaced with alanine residues, and these GST fusion proteins were tested in an in vitro Mek1 kinase assay. These assays showed that the phosphorylation signals of Rdh54-1-T6A and Rdh54-6-T673A, but not of Rdh54-1-T158A, were largely reduced to the level of GST alone, which was used as a negative control (Fig. 6C). These results indicate that the T6 and T673 residues, but not T158, of Rdh54 are phosphorylated by Mek1, at least in vitro.
To investigate whether these phosphorylations depend on Mek1 kinase activity, we constructed a plasmid that harbors a Mek1 kinase-dead (KD) mutant. Then, we prepared extracts of mitotic S. pombe cells that harbor plasmids expressing Mek1-GFP, Mek1-KD-GFP or GFP alone (negative control) under the nmt1 promoter, using Rdh54-1, Rdh54-5/6 and GST alone (negative control) as substrates. Here, we constructed a novel plasmid, Rdh54-5/6, carrying T673 at center, and used it instead of Rdh54-6 because T673 was situated at the N-terminal edge of Rdh54-6 and was easily degraded; this property was incompatible with this experiment. The intact Rdh54-5/6 fragment could be prepared using this new plasmid (arrowhead in Fig. 6D). The phosphorylation levels of Rdh54-1 and Rdh54-5/6 were conspicuously reduced to the level observed in cells transformed with GFP alone, which indicates that Mek1-KD cannot phosphorylate the Rdh54-1 and Rdh54-5/6 fragments, and therefore that the phosphorylation of Rdh54-T6 and Rdh54-T673 depends on Mek1 kinase activity.
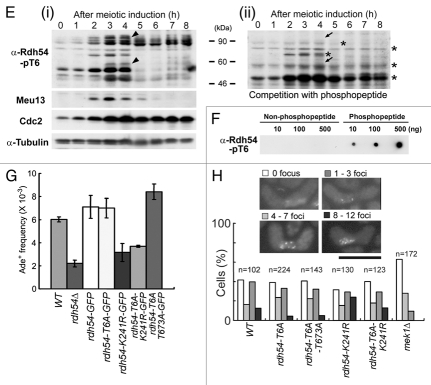
An anti-Rdh54-pT6 antibody was used to examine the in vivo phosphorylation of Rdh54 at T6. This antibody was used in western blot analysis of S. pombe meiotic cell extracts, because Rdh54 is predominantly expressed during early meiosis, forms nuclear foci during the horsetail phase (Sup. Fig. 9), and is required for nuclear division in meiosis I (Sup. Fig. 10). Indeed, the antibody recognized bands of the expected size (∼90 kDa, including Rdh54 degradation products), whose intensities peaked at the time when HR occurred during meiosis (Fig. 6Ei), as judged by the Meu13 peak.40 Cdc2 and α-tubulin were also monitored as a meiotic progression marker and a loading control, respectively. Peptide competition indicated that the bands marked by tilted arrowheads (Fig. 6Ei) are Rdh54-pT6-specific bands recognized by the antibody because they disappeared after the competition experiment (tilted arrows in Fig. 6Eii). A dot blot assay confirmed that the antibody specifically recognized the phospho-T6 peptide (antigen) but not the unphosphorylated T6 peptide (Fig. 6F). These results indicate that Rdh54-T6 is phosphorylated during S. pombe meiosis.
The physiological role of Rdh54 phosphorylations in vivo was explored by measuring gene conversion rates in rdh54-GFP, rdh54-T6A-GFP and rdh54-T6A/T673A-GFP strains. The frequency of meiotic recombination in the rdh54-T6A-GFP strain was almost equal to that of the rdh54-GFP strain, but that of the rdh54-T6A/T673A-GFP double mutant was enhanced by 20% (Fig. 6G). In contrast, the rdh54-K241R-GFP mutant, in which the highly conserved Walker-type A lysine (K) residue in Rdh54 was replaced with arginine (R), and the rdh54Δ mutant showed reduced frequencies of meiotic recombination. The rdh54-T6A/K241R-GFP double mutant strain also showed a similar reduced frequency. Furthermore, the numbers of Rdh54 foci in the rdh54-T6A-GFP and rdh54-T6A/T673A-GFP strains were slightly reduced in the horsetail phase as compared to those of the rdh54-WT-GFP strain (Fig. 6H). Moreover, the Rdh54-T6A/K241R strain had slightly fewer Rdh54 foci than did the rdh54-K241R-GFP strain. In contrast, the number of Rdh54 foci was largely reduced in the mek1Δ strain (leftmost bars in Fig. 6H). Taken together, we conclude that rdh54-T6 and/or rdh54-T673 phosphorylations play a role in meiotic progression.
Discussion
The present study showed that Mek1 has a key role in meiotic recombination by mediating the signal transduction cascade from Rad3 and Tel1 to Mus81 and Rdh54. The regulatory mechanism consists of three steps, as schematically presented in Figure S11 and discussed below.
Rad3 and Tel1 phosphorylate Mek1 to initiate meiotic recombination.
As the first step of Mek1-mediated signal transduction, Rad3 and Tel1 phosphorylate the S12, S14 and T15 residues of Mek1 in vitro (Fig. 1). The presence of shifted bands shown by western blot analysis of S. pombe meiotic cells that express the 9Myc-Mek1 fragment (Fig. 2Bi), and the disappearance of these shifted bands in mek1-100-S12AS14AT15A-9myc mutant cells (Fig. 2Bii), indicate that these phosphorylations occur in vivo during meiosis. We confirmed that these phosphorylations are catalyzed by Rad3 and/or Tel1 in vivo because the uppermost shifted band (white arrowhead in Fig. 2B) was either abolished in rad3Δ and/or tel1Δ cells (Fig. 2C). Notably, the reduction of the second shifted band (black arrowhead in Fig. 2C) was more predominant in tel1Δ cells than that of rad3Δ cells (Fig. 2C), suggesting a more important role of Tel1 than Rad3 in phosphorylation of Mek1. Further, the uppermost shifted band almost disappeared in rec12Δ cells, which cannot produce meiotic DSBs (Fig. 2Di), suggesting that Mek1 phosphorylation is due to programmed DSBs generated by Rec12.
We previously showed that Mek1 is required for the meiotic recombination checkpoint that arrests or delays meiotic cell cycle progression, thereby preventing the formation of defective gametes.23 Here, we show that this may be attained by Mek1 phosphorylation because the mutant strain (mek1-T15A) was substantially delayed in the initiation of meiotic S phase, and this delay was reflected by the prolonged phosphorylation of the Tyr15 residue of Cdc2 (Fig. 3 and Sup. Fig. 5E). Moreover, the mek1-KD (Sup. Fig. 3) and mek1Δ15 mutants did not experience a meiotic delay, while the mek1-FHAD mutant displayed phenotypes intermediate between those of the mek1-T15A and mek1-KD mutants (Sup. Fig. 4). From these results, we conclude that the phosphorylation of Mek1-T15 has an important role in proper meiotic progression, in particular the initiation of meiotic S phase, which may regulate the meiotic recombination checkpoint. This conclusion, however, does not necessarily exclude the possibility that Mek1-T15 may also participate in regulation of early events before DSB formation, which was recently proposed as a novel function of Mek1;15 this will be rigorously examined in our future studies.
Notably, although the recombination frequencies of mek1Δ and mek1-KD cells were reduced, the recombination frequencies of mek1-T15A cells were almost normal, while those of the mek1-FHAD mutant were intermediate between those of the mek1-T15A and mek1Δ mutants (Sup. Fig. 5). Moreover, the addition of the T15A mutation to the mek1-KD and mek1-FHAD alleles had little effect on meiotic recombination frequency, and mek1-S12A, mek1-S14A and mek1-T15A cells also had normal meiotic recombination rates (Sup. Fig. 5D). These results indicate that phosphorylation of S12, S14 and T15 of Mek1 is irrelevant for meiotic recombination function. Moreover, since spore viability is not reduced in any of the Mek1 mutants (Sup. Fig. 5), the kinase activity of Mek1 may play only a minor role in recombination; otherwise spore viability would have to be reduced significantly as it has been demonstrated for many other mutants in S. pombe. Thus, the recombination reductions may be a consequence of the early role of Mek1 in DSB formation as reported by others.15,36
Taken together, we conclude that Mek1 plays multiple roles; phosphorylation of Mek1-T15 is important for proper progression of early meiosis including initiation of meiotic S phase, while the kinase and FHA domains of Mek1 are required for proper meiotic recombination.
Mek1-T318 and -T322 are required for the full activity of Mek1.
In the second step of Mek1-mediated signal transduction, Mek1 phosphorylates itself to enhance the kinase activity that is required for initiating meiotic recombination. Indeed, an in vitro kinase assay first showed that Mek1-T318 is the major autophosphorylation site (Fig. 4C–E). In contrast, in vivo kinase assay using the S. pombe immunoprecipitates (Fig. 4F) and examination of the meiotic recombination rates of S. pombe strains (Fig. 4G) indicated that phosphorylation of both Mek1-T318 and -T322 is required for meiotic recombination. This suggests a sequential activation event; Mek1-T322 participates in the Mek1 activation step after autophosphorylation of Mek1-T318. This hypothesis does not exclude a possibility that activation through autophosphorylation is regulated by an unknown associate of Mek1. Indeed, in mammalian cells, PML (promyelocytic leukemia) interacts with CHK2 and activates it by inducing its autophosphorylation,41 which is an essential step for CHK2 activity that occurs after phosphorylation by the upstream kinase ATM (ataxia telangiectasia-mutated).
Phosphorylation of both T354 and T358 in the activation domain of S. cerevisiae Rad53 (a Chk2 homolog), which correspond to T318 and T322 of S. pombe Mek1, is essential for activation of its kinase activity, not only upon DNA damage but also during the normal cell cycle, and mutation of these residues caused defects in the checkpoint response and hyper-sensitivity to DNA damage and DNA replication stress agents.42 The dimeric crystal structure of the kinase-inactive mutant of human CHK2 revealed that dimerization promotes kinase activation through activation-loop autophosphorylation, and the productive CHK2 dimerization additionally involves the intermolecular FHA-kinase domain and FHA-FHA interactions.41 Dimerization serves to optimally position the kinase active sites for efficient activation loop transphosphorylation by positioning the kinase active sites so that they closely face each other. If the Mek1 structure is similar to that of CHK2, phosphorylation of both T318 and T322 of Mek1 may also play pivotal roles for its activation.
Downstream phosphorylation targets of Mek1 during meiosis.
As the third step of Mek1-mediated signal transduction, we identified the phosphorylation targets of Mek1 that regulate meiotic progression, and in particular meiotic recombination. We first showed that the immunoprecipitates of Cds1/Mek1 phosphorylates Mus81 on T218, T275 and T422 (Figs. 5 and Sup. Fig. 6). It was previously shown that Mus81 is phosphorylated by an unknown kinase on T239 (now T275) in the T-X-X-D motif in response to HU-induced replication arrest.25,32 Here, we referred to their T239 as T275, according to the revised DNA sequence in the S. pombe database (http://www.genedb.org/genedb/Search?name=SPCC4G3.05c&organism=pombe). We propose here that this unknown kinase is most probably Cds1 and/or Mek1, because either could directly phosphorylate Mus81 on T275 in vitro (Sup. Fig. 6F). Failure of Mus81 phosphorylation by the Mek1-KD form also supports this proposal (Fig. 5A). Since the FHA domain is a phosphopeptide-binding module with a strong preference for the T-X-X-D motif, it was proposed that T275 phosphorylation of the Mus81 T-X-X-D motif may promote the interaction of Mus81 with the FHA domain of Cds1. This interaction, which removes Mus81 from chromatin, thus negatively regulates the Mus81 function in DNA cleavage.32 Moreover, the recombination frequency in mus81.T239A cells after acute exposure to HU was more enhanced than in WT cells,32 which could also happen during meiosis. Since Mus81-T275 negatively regulates DNA cleavage by removing Mus81 from chromatin, and the T275A mutation caused abnormal recombination,32 our results (Sup. Fig. 5) does not directly link the phosphorylation signal cascade originating with Tel1/Rad3 and terminating with meiotic recombination; this is probably due to Mus81 phosphorylation primarily by Cds1 during meiosis. The physiological significance of the Mus81-T218 and -T422 phosphorylations remains unclear, due to the already reduced recombination rates of the corresponding mutants (Sup. Fig. 8).
Unlike Mek1, the phosphorylation targets of Cds1 in mitosis are well known. For example, Cds1 phosphorylates the S32, S34 and T72 residues of an essential nuclear protein, Rad60, which is involved in repairing DSBs.44 Rad60-pT72 mediates the Cds1-Rad60 interaction in response to replication arrest, whereas Rad60-pS32 and -pS34 are critical for surviving replication stress.44 Rad60 phosphorylation is also required to regulate HR at stalled replication forks by ensuring the structural maintenance of the protein complex of chromosomes 5 and 6 (Smc5/6).45 Nrm1, a transcriptional repressor that acts at the G1 to S phase transition, is phosphorylated by Cds1 in response to DNA replication stress, which leads to the expression of genes important for DNA replication and repair.46 Cdc25, a well known Cds1 target, is also phosphorylated by Mek1, which also regulates the nuclear localization of Cdc25 and cell cycle arrest.47
As a novel target, we also found that Mek1 phosphorylates Rdh54 at T6 and T673 in vitro (Fig.6A–D). Phosphorylation of Rdh54-T6 occurs in vivo because an anti-Rdh54-pT6 antibody recognized bands of the expected size for Rdh54 that peaked at the time of meiotic HR, as shown by western blot analysis (Fig. 6Ei). The identification of these bands as Rdh54-pT6 was confirmed by competition experiments and dot blot analysis (Figs. 6Eii and F). Since the HR frequency of Rdh54-T6A/T673A mutant cells was enhanced (Fig. 6G), and the extent of abnormal chromosome segregation and the number of Rdh54 foci were reduced in Rdh54-T6A cells (Fig. 6H), phosphorylation of the T6 and T673 residues may play a role in meiotic progression.
We also showed that Mek1 phosphorylates Rhp54 and Mcp6 (Fig. 6B), a meiosis-specific protein that is required for astral microtubule positioning to maintain horsetail movement in meiosis I.38,39 Determination of the phosphorylation sites of Rhp54 and Mcp6 and other Mek1 association partners, and of their physiological significance, is the target of future experiments.
Experimental Procedures
Synchronous meiosis.
Fresh homozygous pat1-114 diploid strains were grown in EMM with supplements at 25°C for >24 h. Cells at the mid-log phase were collected, washed and then transferred at a density of 2 × 106 cells/mL to EMM2 supplemented with leucine (62.5 µg/mL) and uracil (18.8 µg/mL), but lacking NH4Cl. After 16 h of incubation at 25°C, NH4Cl (5 mg/mL), leucine (187.5 µg/mL) and uracil (56.3 µg/mL) were added to the culture medium, and the temperature was raised to 34°C to induce meiosis. Meiotic progression was monitored by counting the nuclei of cells fixed in 70% ethanol and stained with Hoechst33342 (Wako Co., Ltd., Osaka, Japan).
Protein extraction and western blotting.
Protein extracts were prepared as previously described in reference 36. For Mek1-9myc detection, the blots were probed with the mouse monoclonal antibody 9E10 (Upstate Biotechnology) and, as a loading control, α-tubulin was detected with an anti-tubulin antibody (Sigma T5168). For detecting Cdc2 Tyr15 phosphorylation, the blots were probed with an anti-phospho-Cdc2 (Tyr15) rabbit polyclonal antibody (NEB 9111). To detect Cdc2 as a loading control, the anti-PSTAIRE antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used.
Recombination frequency and spore viability.
Crossing-over rates were determined as previously described in reference 23 and 38. Briefly, haploid parental strains were grown on YPD plates at 30°C, and cells were mated and sporulated on ME plates at 28°C (zygotic meiosis). After 1 d of incubation, the spores were isolated. The frequency of crossing-over was determined by measuring the genetic distance between his2+ and leu1+. The gene conversion rate was determined as previously described in reference 23. For this, haploid parental strains were grown on YPD at 33°C. Cells were mated and sporulated on ME plates at 28°C (zygotic meiosis). After 2 d of incubation, the spores were treated with 1% glusulase (NEN Life Sciences, Perkin-Elmer, Boston, MA) for 2–3 h at room temperature and checked under a microscope for the complete digestion of contaminating vegetative cells. The glusulase-treated spores were washed with water, and the gene conversion rate and spore viability were measured. The frequency of gene conversion was determined with the ade6-M26 and ade6-469 alleles, as gene conversion between these alleles produces the ade6+ allele.
Mek1 and Cds1 kinase assay in vitro.
Logarithmically growing cells (approximately 5 × 106 cells/mL) that express the Mek1-9Myc or Mek1-KD-9Myc proteins from the nmt41 promoter (pREP41) or Cds1-2HA/6His proteins from the native cds1+ promoter were harvested. Cell extracts were obtained by disrupting the cells with glass beads in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% glycerol, 1% NP-40, 50 mM NaF, 1 mM Na3VO4, 100 µg/mL PMSF, 1 µg/mL aprotinin, 1 µg/mL pepstatin, 1 µg/mL leupeptin, 1 mM DTT) and centrifuging the extract at 15,000 rpm for 15 min at 4°C. Mek1-9Myc and Cds1-2HA/6His were immunoprecipitated with the mouse monoclonal antibody 9E10 (Upstate Biotechnology, Charlottesville, VA), the mouse monoclonal antibody PL14 (MBL, Nagoya, Japan) or the rat monoclonal antibody 3F10 (Roche, Basel, Switzerland). In vitro kinase assays were performed in kinase buffer (25 mM HEPES-KOH [pH 7.5], 50 mM KCl, 10 mM MgCl2, 10 mM MnCl2, 2% glycerol, 0.1% NP-40, 1 mM Na3VO4, 1 mM DTT). Kinase activity was measured by adding 5 µL purified Mek1-9myc or Cds1-2HA/6His to 25 µL of a reaction mixture containing 10 µM ATP, [γ-32P] ATP and 15 µL substrate. The reaction was conducted at 30°C for 30 min and then stopped by the addition of 10 µL 4x SDS sample buffer and boiled. Samples were subjected to SDS-PAGE (10% gel) and detected using the Bio-imaging Analyzer BAS 1800II (Fuji Film, Tokyo, Japan).
Acknowledgements
We are indebted to Professors C. Shimoda, M. Yamamoto, T. Nakamura, Y. Watanabe, S.L. Forsburg, J. Kohli and the National BioResource Project (http://yeast.lab.nig.ac.jp/nig/) for S. pombe strains. We also thank Ms. T. Motoyama and Ms. E. Okamura for technical assistance and Dr. K. Smith of Bioedit Ltd., for critically reading the manuscript. This work was supported in part by grants-in-aid for Scientific Research on Priority Areas “Applied Genomics”, Scientific Research (S), Exploratory Research, and the Science and Technology Incubation Program in Advanced Regions from the Ministry of Education, Culture, Sports, Science and Technology of Japan to Hiroshi Nojima, and by NIH grant GM59447 to Paul Russell.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/14050
Supplementary Material
References
- 1.Ciccia A, McDonald N, West SC. Structural and Functional Relationships of the XPF/MUS81 Family of Proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 2.Smith GR. Genetic analysis of meiotic recombination in Schizosaccharomyces pombe. Methods Mol Biol. 2009;557:65–76. doi: 10.1007/978-1-59745-527-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huertas P. DNA resection in eukaryotes: Deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartagena-Lirola H, Guerini I, Manfrini N, Lucchini G, Longhese MP. Roles of the S. cerevisiae Rad53 checkpoint kinase in signalling double-strand breaks during the meiotic cell cycle. Mol Cell Biol. 2008;28:4480–4493. doi: 10.1128/MCB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res. 2009;19:2245–2257. doi: 10.1101/gr.096297.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi N, Barot A, Jamison C, Börner GV. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009;5:1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin FM, Lai YJ, Shen HJ, Cheng YH, Wang TF. Yeast axial-element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 2010;29:586–596. doi: 10.1038/emboj.2009.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailis JM, Roeder GS. Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell. 2000;101:211–221. doi: 10.1016/S0092-8674(00)80831-4. [DOI] [PubMed] [Google Scholar]
- 10.Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol Biol Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, et al. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 13.Cromie G, Smith GR. Meiotic Recombination in Schizosaccharomyces pombe: A Paradigm for Genetic and Molecular Analysis. In: Egel R, Lankenau DH, editors. Recombination and Meiosis. Vol. 3. Berlin: Springer; 2007. pp. 196–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;27:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latypov V, Rothenberg M, Lorenz A, Octobre G, Csutak O, Lehmann E, et al. The roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination-partner choice in Schizosaccharomyces pombe. Mol Cell Biol. 2010;30:1570–1581. doi: 10.1128/MCB.00919-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhind N, Russell P. Chk1 and Cds1: Linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branzei D, Foiani M. The Rad53 signal transduction pathway: Replication fork stabilization, DNA repair and adaptation. Exp Cell Res. 2006;312:2654–269. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Poon RY. The multiple checkpoint functions of CHK1 and CHK2 in maintenance of genome stability. Front Biosci. 2008;13:5016–5029. doi: 10.2741/3060. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka K, Russell P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat Cell Biol. 2001;3:966–972. doi: 10.1038/ncb1101-966. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka K, Russell P. Cds1 phosphorylation by Rad3-Rad26 kinase is mediated by forkhead-associated domain interaction with Mrc1. J Biol Chem. 2004;279:32079–32086. doi: 10.1074/jbc.M404834200. [DOI] [PubMed] [Google Scholar]
- 21.Xu YJ, Davenport M, Kelly TJ. Two-stage mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes Dev. 2006;20:990–1003. doi: 10.1101/gad.1406706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino K, Masai H. Rad3-Cds1 mediates coupling of initiation of meiotic recombination with DNA replication. Mei4-dependent transcription as a potential target of meiotic checkpoint. J Biol Chem. 2006;281:1338–1344. doi: 10.1074/jbc.M505767200. [DOI] [PubMed] [Google Scholar]
- 23.Shimada M, Nabeshima K, Tougan T, Nojima H. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 2002;21:2807–2818. doi: 10.1093/emboj/21.11.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez-Hidalgo L, Moreno S, San-Segundo PA. Regulation of meiotic progression by the meiosis-specific checkpoint kinase Mek1 in fission yeast. J Cell Sci. 2003;116:259–271. doi: 10.1242/jcs.00232. [DOI] [PubMed] [Google Scholar]
- 25.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 27.Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81. Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;65:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holzen TM, Shah PP, Olivares HA, Bishop DK. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 2006;20:2593–604. doi: 10.1101/gad.1447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nimonkar AV, Amitani I, Baskin RJ, Kowalczykowski SC. Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J Biol Che. 2007;282:30776–30784. doi: 10.1074/jbc.M704767200. [DOI] [PubMed] [Google Scholar]
- 30.Catlett MG, Forsburg SL. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol Biol Cell. 2003;14:4707–4720. doi: 10.1091/mbc.E03-05-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi P, Kwon Y, Moses DN, Seong C, Sehorn MG, Singh AK, et al. Functional interactions of meiotic recombination factors Rdh54 and Dmc1. DNA Repair (Amst) 2009;8:279–284. doi: 10.1016/j.dnarep.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kai M, Boddy MN, Russell P, Wang TS. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 2005;19:919–932. doi: 10.1101/gad.1304305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 34.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol. 2009;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothenberg M, Kohli J, Ludin K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 2009;5:1000722. doi: 10.1371/journal.pgen.1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahn J, Urist M, Prives C. The Chk2 protein kinase. DNA Repair (Amst) 2004;3:1039–1047. doi: 10.1016/j.dnarep.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 38.Saito TT, Tougan T, Kasama T, Okuzaki D, Nojima H. Mcp7, a meiosis-specific coiled-coil protein of fission yeast, associates with Meu13 and is required for meiotic recombination. Nucleic Acids Res. 2004;32:3325–3339. doi: 10.1093/nar/gkh654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka K, Kohda T, Yamashita A, Nonaka N, Yamamoto M. Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr Biol. 2005;15:1479–1486. doi: 10.1016/j.cub.2005.07.058. [DOI] [PubMed] [Google Scholar]
- 40.Nabeshima K, Kakihara Y, Hiraoka Y, Nojima H. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 2001;20:3871–3881. doi: 10.1093/emboj/20.14.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S, Jeong JH, Brown AL, Lee CH, Pandolfi PP, Chung JH, et al. Promyelocytic leukemia activates Chk2 by mediating Chk2 autophosphorylation. J Biol Chem. 2006;281:26645–26654. doi: 10.1074/jbc.M604391200. [DOI] [PubMed] [Google Scholar]
- 42.Fiorani S, Mimun G, Caleca L, Piccini D, Pellicioli A. Characterization of the activation domain of the Rad53 checkpoint kinase. Cell Cycle. 2008;7:493–499. doi: 10.4161/cc.7.4.5323. [DOI] [PubMed] [Google Scholar]
- 43.Cai Z, Chehab NH, Pavletich NP. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol Cell. 2009;35:818–829. doi: 10.1016/j.molcel.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Raffa GD, Wohlschlegel J, Yates JR, 3rd, Boddy MN. SUMO-binding motifs mediate the Rad60-dependent response to replicative stress and self-association. J Biol Chem. 2006;281:27973–27981. doi: 10.1074/jbc.M601943200. [DOI] [PubMed] [Google Scholar]
- 45.Miyabe I, Morishita T, Shinagawa H, Carr AM. Schizosaccharomyces pombe Cds1Chk2 regulates homologous recombination at stalled replication forks through the phosphorylation of recombination protein Rad60. J Cell Sci. 2009;122:3638–3643. doi: 10.1242/jcs.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Bruin RA, Kalashnikova TI, Aslanian A, Wohlschlegel J, Chahwan C, Yates JR, 3rd, et al. DNA replication checkpoint promotes G1-S transcription by inactivating the MBF repressor Nrm1. Proc Natl Acad Sci USA. 2008;105:11230–11235. doi: 10.1073/pnas.0801106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pérez-Hidalgo L, Moreno S, San-Segundo PA. The fission yeast meiotic checkpoint kinase Mek1 regulates nuclear localization of Cdc25 by phosphorylation. Cell Cycle. 2008;7:3720–3730. doi: 10.4161/cc.7.23.7177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.