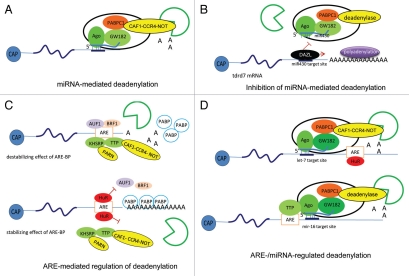
Figure 4.
Models of multicomponent complexes required for regulation of polyadenylation/deadenylation processes. (A) miRNA-mediated deadenylation. miRISCs, which contain Ago, GW182, PABPC1 and either CAF1-CCR4-NOT (as indicated) or Pan2-Pan3 (not shown) deadenylases, deliver miRNAs to the target mRNAs and mediate deadenylation, which leads subsequently to mRNA degradation. (B) Inhibition of miRNA-mediated deadenylation. The RNA-binding protein DAZL overcomes the inhibitory effect of miRNA by inducing polyadenylation and leading to active translation even in the presence of miRNA. (C) ARE-mediated regulation of deadenylation. The ARE-binding proteins (BP) mediate destabilization and/or stabilization of the ARE-containing mRNAs. ARE-BPs, such as AUF1, TTP, BRF1 and KHSRP, recruit deadenylases, such as PARN and CAF1-CCR4-NOT, to target ARE-mRNAs and initiate the deadenylation process that precedes degradation. Another ARE-BP, HuR, plays a role in stabilizing ARE-containing mRNAs by blocking the binding of ARE-BPs involved in the destabilization of ARE-mRNAs, such as AUF1, TTP and KHSRP. This competition stabilizes the association of PABP to the poly(A) tail or prevents the recruitment of deadenylases to the ARE-mRNA. (D) ARE- and miRNA-regulated deadenylation. Cooperation of AREs-BPs, miRNAs, deadenylases and exosome is essential for the regulation of mRNA stability. The recruitment of the ARE-BPs HuR or TTP to the ARE sequence assists the targeting of let-7- or miR-16-loaded miRISC complexes, respectively, to the most proximal site to the ARE sequence.