Abstract
Homologous recombination is needed for meiotic chromosome segregation, genome maintenance, and tumor suppression. RAD51AP1 (RAD51 associated protein 1) has been shown to interact with and enhance the recombinase activity of RAD51. Accordingly, genetic ablation of RAD51AP1 leads to enhanced sensitivity to and also chromosome aberrations upon DNA damage, demonstrating a role for RAD51AP1 in mitotic homologous recombination. Here we show physical association of RAD51AP1 with the meiosis-specific recombinase DMC1 and a stimulatory effect of RAD51AP1 on the DMC1-mediated D-loop reaction. Mechanistic studies have revealed that RAD51AP1 enhances the ability of the DMC1 presynaptic filament to capture the duplex-DNA partner and to assemble the synaptic complex, in which the recombining DNA strands are homologously aligned. We also provide evidence that functional cooperation is dependent on complex formation between DMC1 and RAD51AP1 and that distinct epitopes in RAD51AP1 mediate interactions with RAD51 and DMC1. Finally, we show that RAD51AP1 is expressed in mouse testes, and that RAD51AP1 foci colocalize with a subset of DMC1 foci in spermatocytes. These results suggest that RAD51AP1 also serves an important role in meiotic homologous recombination.
Keywords: DNA repair, genomic stability, meiotic proteins
Homologous recombination (HR) is an important pathway for the elimination of DNA double-strand breaks (DSBs) and other deleterious lesions from chromosomes (1). Defects in HR lead to destabilization of the genome and represent a progenitor of the tumor phenotype in mammals. Moreover, HR in meiosis generates crossovers between homologous chromosomes that are essential for chromosome segregation during meiosis I (1, 2). Accordingly, HR mutants often exhibit severe meiotic phenotypes, including prophase arrest or apoptosis (2).
The eukaryotic HR reaction is mediated by RAD51 or DMC1, recombinases that are orthologous to bacterial RecA (1–3). Like RecA, RAD51 and DMC1 function within the context of a right-handed helical polymer assembled on ssDNA. This recombinase-ssDNA nucleoprotein filament, commonly referred to as the presynaptic filament, possesses a second binding site that can accommodate a duplex-DNA molecule. As such, the presynaptic filament provides the catalytic scaffold to sample the incoming duplex DNA for homology and form first an unstable paranemic joint with the duplex, which can mature into a stable plectonemic joint called D loop, in which the recombining DNA strands are linked topologically (4, 5).
A variety of recombinase partner proteins that have specific attributes germane for the enhancement of the HR reaction have been identified (5). RAD51AP1 (RAD51 associated protein 1), so named because of its ability to interact with human RAD51 (6), is a vertebrate-specific protein that enhances the ability of RAD51 to mediate the D-loop reaction (7, 8). RAD51AP1 knockdown by RNA interference leads to cellular sensitivity to DNA damaging agents and elevated DNA damage-induced chromatid breaks (7, 8). Enhancement of the RAD51 recombinase activity by RAD51AP1 is contingent upon physical interaction of the two HR factors (7, 8). It is not clear whether RAD51AP1 is involved in meiotic recombination, and, if so, whether it functions with RAD51, DMC1, or both recombinases.
Here, we document studies that reveal physical interaction of RAD51AP1 with the meiotic recombinase DMC1 and strong enhancement of the DMC1-mediated D-loop reaction by RAD51AP1. We provide evidence that RAD51AP1 cooperates with the DMC1 presynaptic filament to capture duplex DNA and to assemble the synaptic complex, in which the recombining DNA molecules are joined paranemically. Moreover, using spliced forms and mutant variants of RAD51AP1, we show that physical interaction of the RAD51AP1-DMC1 pair is indispensable for their functional synergy. We present evidence that RAD51AP1 is abundantly expressed in mouse meiosis and that RAD51AP1 colocalizes with DMC1 in foci on meiotic chromatin. Our findings provide insights into the mechanistic role of RAD51AP1 in HR reactions and suggest that RAD51AP1 is also a regulator of meiotic HR.
Results
Purification of RAD51AP1 Isoforms.
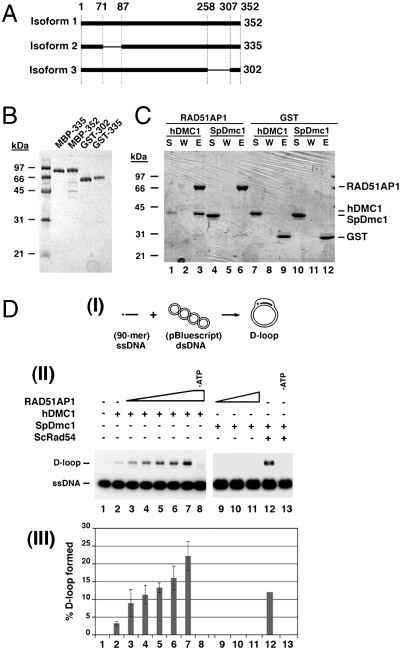
Three RAD51AP1 isoforms that harbor 352 residues (isoform 1), 335 residues (isoform 2), or 302 residues (isoform 3) were used in the present work (Fig. 1A). Isoform 2 was used for initial studies with DMC1 as it is the predominant species in humans and the only documented isoform in some species (Table S1). Published procedures (8) were used to purify these RAD51AP1 isoforms (Fig. 1B).
Fig. 1.
Characterization of RAD51AP1 isoforms. (A) The three human RAD51AP1 isoforms used in this study. (B) SDS-PAGE analysis of the purified tagged RAD51AP1 isoforms. (C) In a pull-down assay, GST-tagged RAD51AP1 Isoform 2 was found to associate with human DMC1 (lane 3) but not with S. pombe Dmc1 (SpDmc1; lane 6). Control reactions were carried out with GST. The supernatant (S), wash (W), and SDS eluate (E) of the pull-down reactions were analyzed by SDS-PAGE. (D) RAD51AP1 isoform 2 specifically stimulates D-loop formation by human DMC1. (I) Schematic of the D-loop assay. (II) RAD51AP1 (50, 100, 200, 300, or 500 nM) enhanced the D-loop reaction mediated by human DMC1 (lanes 3 to 7) in an ATP-dependent fashion (lane 8). SpDmc1 was refractory to RAD51AP1 (200, 300, or 500 nM; lanes 9 to 11) but was enhanced by ScRad54 (200 nM; lane 12) in an ATP-dependent fashion (lane 13). (III) The results were plotted; error bars represent the standard deviation (± SD) calculated based on at least three independent experiments.
Complex of RAD51AP1 and DMC1.
An affinity pull-down assay was employed to examine the ability of GST-tagged RAD51AP1 isoform 2 to interact with human DMC1. As shown in Fig. 1C, a RAD51AP1-DMC1 complex could be readily captured, but little if any of Schizosaccharomyces pombe Dmc1 (SpDmc1) associated with RAD51AP1 (Fig. 1C). We note that neither Saccharomyces cerevisiae Rad51 (yRad51) nor RecA interacts with RAD51AP1 either (8). Maltose binding protein (MBP)-tagged RAD51AP1 isoform 2 is also capable of DMC1 interaction (see below). These results revealed that RAD51AP1 physically interacts with DMC1.
Enhancement of DMC1-Mediated D-Loop Formation by RAD51AP1.
We asked whether RAD51AP1 would enhance the DMC1-mediated D-loop reaction. Indeed, although DMC1 alone converted ∼3% of the input oligonucleotide into the D-loop product (Fig. 1D), up to an 8-fold increase of product formation occurred when RAD51AP1 isoform 2 was added (Fig. 1D). The stimulatory effect of RAD51AP1 on DMC1 is specific, as no such enhancement was observed with SpDmc1 (Fig. 1D). As expected, the D-loop reaction mediated by the combination of DMC1 and RAD51AP1 was ATP dependent (Fig. 1D). Taken together, the results revealed an ability of RAD51AP1 to up-regulate the recombinase activity of DMC1.
The above D-loop experiments were conducted with Mg2+ ion wherein turnover of the recombinase from ssDNA occurred upon ATP hydrolysis. Importantly, RAD51AP1 also stimulated D-loop formation when Mg2+ was substituted by Ca2+ (Fig. S1A) to stabilize the presynaptic filament (9). These results, coupled with the fact that RAD51AP1 binds dsDNA preferentially over ssDNA (8), indicated that RAD51AP1 acts at a step subsequent to the assembly of the presynaptic filament, a premise that will be elaborated upon below.
Promotion of Duplex Capture and Synaptic Complex Assembly by RAD51AP1.
During the homologous pairing process that results in D-loop formation, the presynaptic filament first engages the dsDNA partner and then conducts a search to locate homology in the partner. This leads to the assembly of a nucleoprotein ensemble termed the synaptic complex, in which the recombining ssDNA and dsDNA are homologously aligned (4, 5, 10). We queried whether RAD51AP1 functions with the DMC1 presynaptic filament to engage duplex DNA and promote synaptic complex assembly.
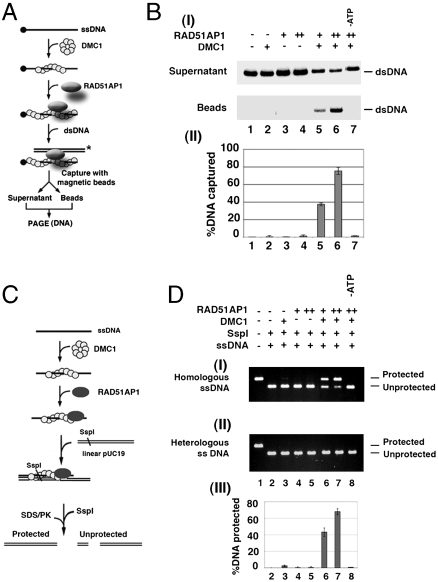
We first tested the effect of RAD51AP1 on duplex-DNA capture, using our previously published procedure (11). For this, the DMC1 presynaptic filament was assembled on ssDNA linked to magnetic beads via a streptavidin–biotin bridge. Then, the ability of the presynaptic filament to capture a radiolabeled duplex DNA of sequence unrelated to the bound ssDNA was examined, either in the absence or presence of RAD51AP1 (see Fig. 2A). In agreement with previously published results (12), the DMC1 presynaptic filament on its own had only a limited ability to capture duplex DNA (Fig. 2B). Importantly, RAD51AP1 strongly enhanced duplex capture by the DMC1 presynaptic filament (Fig. 2B). The duplex capture reaction showed a strict dependence on ATP (Fig. 2B) and DNA (Fig. S1B). Interestingly, the combination of the DMC1 presynaptic filament and RAD51AP1 was much less adept at capturing ssDNA (Fig. S1C) consistent with the dsDNA binding preference of RAD51AP1 (8). Enhancement of duplex capture by RAD51AP1 is specific to human DMC1, as no such effect was seen with SpDmc1 (Fig. S1E). In addition, the DMC1 presynaptic filament stabilized by Ca2+ ion (9) was also unable to capture duplex DNA unless RAD51AP1 was included (Fig. S1D).
Fig. 2.
Functional synergy of RAD51AP1 and DMC1 in duplex capture and synaptic complex assembly. (A) Schematic of the duplex capture reaction. (B) DMC1 alone (lane 2) or RAD51AP1 alone (lanes 3 and 4) exhibited little duplex capture activity. DMC1 synergized with RAD51AP1 (300 or 600 nM) in duplex-DNA capture (lanes 5 and 6), with a dependence on ATP (lane 7). The percentages of captured DNA were quantified and plotted in (II). Error bars represent ± SD calculated based on at least three independent experiments. (C) The basis for the protection of the target DNA against digestion by the restriction enzyme SspI is illustrated. (D) RAD51AP1 (0.5 or 1 μM) was incubated with the DMC1 presynaptic filament assembled on either the homologous ssDNA (I) or heterologous ssDNA (II), and the protection of the dsDNA against digest by Ssp1 was assessed (lanes 6 and 7). Various controls (lanes 2–5) were included. DNA protection was contingent upon ATP being present (lane 8). The results were plotted. Error bars represent ± SD calculated based on at least three independent experiments.
We next assessed synaptic complex formation (12, 13) by DMC1 and RAD51AP1. This assay involves monitoring the protection of linear dsDNA by a DMC1 presynaptic filament harboring homologous ssDNA against digestion by the restriction enzyme SspI (Fig. 2C). As expected from published work (12), the DMC1 presynaptic filament alone was poorly adept at assembling the synaptic complex (Fig. 2D). Importantly, the addition of RAD51AP1 afforded a marked stimulation of synaptic complex assembly, in a manner that required ATP (Fig. 2D). Control reactions revealed that RAD51AP1 does not synergize with the SpDmc1 presynaptic filament in synaptic complex assembly (Fig. S1F).
Requirement for DMC1-RAD51AP1 Interaction in Synaptic Complex Assembly.
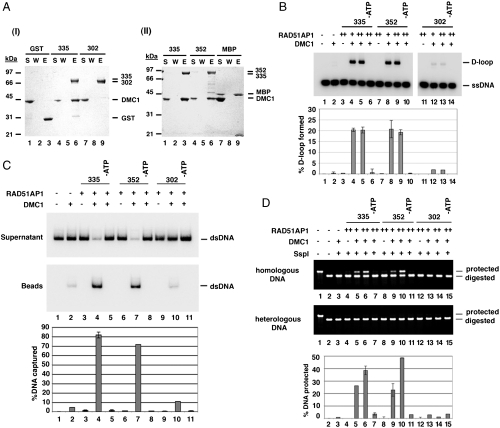
We verified that RAD51AP1 isoform 1 (7) is just as capable as RAD51AP1 isoform 2 in DMC1 interaction (Fig. 3A) and in enhancement of the D-loop reaction, duplex capture, and synaptic complex assembly (Fig. 3 B–D). Interestingly, we found that RAD51AP1 isoform 3, which lacks a segment of the C terminus of the other two isoforms (Fig. 1A), although capable of RAD51 interaction (Fig. S2A), is deficient in DMC1 interaction (Fig. 3A). Accordingly, even though RAD51AP1 isoform 3 was just as active as the other two isoforms in enhancing RAD51’s recombinase activity (Fig. S2B) and in DNA binding (Fig. S2 C–E), it failed to synergize with the DMC1 presynaptic filament in the D-loop reaction or in duplex capture and synaptic complex assembly (Fig. 3 B–D).
Fig. 3.
Lack of physical or functional interaction of RAD51AP1 Isoform 3 with DMC1. (A) Affinity pull-down analyses using GST-tagged RAD51AP1 (I) and MBP-tagged RAD51AP1 (II) isoforms revealed that isoform 3 does not interact with DMC1 (I, lane 9). Control pull-down reactions using GST and MBP were also included. The supernatant (S), wash (W), and SDS eluate (E) fractions of the pull-down reactions were analyzed by SDS-PAGE. (B) Although RAD51AP1 isoform 1 (lanes 8 and 9) and isoform 2 (lanes 4 and 5) were equally capable of enhancing the DMC1-mediated D-loop reaction in an ATP-dependent manner (lanes 6 and 10), RAD51AP1 isoform 3 (lanes 12 and 13) was devoid of such ability. The amount of each RAD51AP1 isoform used was 400 or 600 nM. The percentages of D-loop products formed were quantified and plotted. (C) Isoform 1 and isoform 2 were both found capable of functional synergy with DMC1 in duplex-DNA capture (lanes 4 and 7) in an ATP-dependent fashion (lanes 5 and 8); isoform 3 was much less effective (lane 10). Various controls were included. The amount of each RAD51AP1 isoform used was 600 nM. The results were plotted. (D) Although RAD51AP1 isoform 1 and isoform 2 were both capable of functional synergy with DMC1 in synaptic complex assembly (lanes 5, 6, 9, and 10) in an ATP-dependent fashion (lanes 7 and 11), isoform 3 was much less effective (lanes 13 and 14). Various controls were included. The amount of MBP-tagged RAD51AP1 isoform 1 and GST-tagged RAD51AP1 isoform 2 or isoform 3 used was 300 or 500 nM. The results were plotted. (B–D) Error bars represent the standard deviation (± SD) calculated based on at least three independent experiments.
RAD51AP1 Mutants Defective in RAD51 Association Are Proficient in DMC1 Interactions.
The L319Q and H329A (14) mutants of RAD51AP1 isoform 2 are deficient in physical and functional interactions with RAD51 (8). Interestingly, these RAD51AP1 mutant proteins (Fig. S3A) are just as proficient as the wild-type counterpart in DMC1 interaction (Fig. S3B). When tested in the D-loop, duplex capture, and synaptic complex assembly assays, the two mutants showed a stimulatory activity comparable to that of RAD51AP1 isoform 1 or isoform 2 (Fig. S3 C–E). Overall, these results revealed that RAD51AP1 residues that are critical for RAD51 interaction are, in fact, dispensable for physical association and functional synergy with DMC1.
Meiotic Expression of RAD51AP1.
We conducted RT-PCR with total RNA isolated from testes of juvenile mice [5, 15, and 21 d postpartum (dpp); Fig. S4B], with the three different age samples being enriched for premeiotic, midprophase I, and late prophase I meiotic cells (15). Even though three RAD51AP1 isoforms exist in human cells, only one isoform (Fig. S4A), equivalent to the human isoform 2, has been documented in mice (Table S1; 16). RT-PCR and immunoblot analyses revealed that mouse RAD51AP1 is abundantly expressed in 15 and 21 dpp testes (Fig. S4 B–C). Our results are in concordance with previous reports (14, 16) showing the presence of the RAD51AP1 transcript in human or mouse testes.
Colocalization of RAD51AP1 with DMC1 on Meiotic Chromatin.
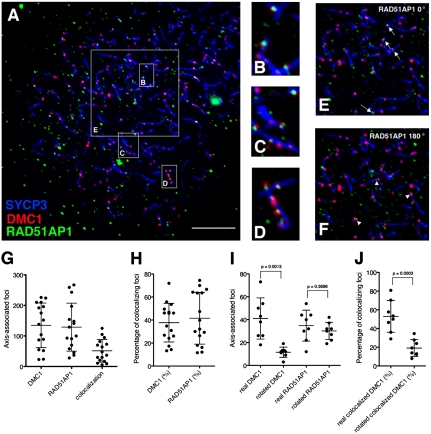
Next we wanted to ask if RAD51AP1 colocalizes with DMC1, which appears as discrete foci on chromatin during the leptotene and zygotene stages (17). Meiotic chromosome spreads were prepared from spermatocytes of juvenile mice and triple-stained with antibodies raised against SYCP3, DMC1, and RAD51AP1. SYCP3 is a component of meiotic chromosome axes (18, 19) used to monitor meiotic progression (20). Several classes of RAD51AP1 foci could be discerned (Fig. S5 and companion description online); below, we focus on axis-associated class I foci. Among 16 leptotene spermatocytes analyzed (Fig. 4A), frequent colocalization of RAD51AP1 with DMC1 was observed (Fig. 4 B–D). These nuclei had 135 ± 73 DMC1 foci and 129 ± 79 class I RAD51AP1 foci, of which 51 ± 37 foci stained with antibodies to both proteins (Fig. 4G). The fraction of DMC1 foci positive for RAD51AP1 was 38 ± 17%, and the fraction of RAD51AP1 foci positive for DMC1 was 42 ± 23% (Fig. 4H).
Fig. 4.
RAD51AP1 colocalizes with DMC1 foci on meiotic chromatin. (A) Example of a spermatocyte nucleus in leptonema. Chromosome spreads were stained with anti-SYCP3 (blue), anti-DMC1 (red), and anti-RAD51AP1 GP49 (green) antibodies. Scale bar = 10 μm. (B–D) Zoomed-in displays of regions indicated in A that harbored colocalized foci of RAD51AP1 and DMC1. Note that foci colocalization was scored without restriction to the subset of foci that share the same centroid. (E and F) Rotation experiment evaluating the statistical significance of RAD51AP1-DMC1 colocalization. E is a zoomed-in display of the 20 × 20 μm box marked in A within which colocalized RAD51AP1 and DMC1 foci were counted, as indicated by white arrows. F corresponds to the same region in E but with the RAD51AP1 immunofluorescence channel rotated 180°. RAD51AP1 foci that overlap with DMC1 foci after image rotation, as indicated by white arrowheads, are considered fortuitously colocalized. (G) Counts of axis-associated DMC1, RAD51AP1, and colocalized foci in leptotene stage spermatocytes. Each dot represents the count from one nucleus and the same 16 nuclei were quantified (bars = mean ± SD). (H) Colocalized foci as a percentage of DMC1 or RAD51AP1 total foci. Each dot represents the percentage from one nucleus and 16 nuclei were quantified (bars = mean ± SD). (I) Results of a rotation experiment wherein axis-associated DMC1 or RAD51AP1 foci were counted before and after 180° rotation of the DMC1 or RAD51AP1 immunofluorescence channel. Each dot represents the count from one nucleus and eight nuclei were quantified (bars = mean ± SD). (J) Colocalized foci as a percentage of DMC1 total foci before and after 180° rotation of the RAD51AP1 immunofluorescence channel. Each dot represents the count from one nucleus and eight nuclei were quantified (bars = mean ± SD).
We tested whether the colocalization of RAD51AP1 with either SYCP3-stained axes or DMC1 foci could have arisen from fortuitous spatial proximity of unrelated protein distributions (21, 22). First, we counted colocalizing RAD51AP1 and DMC1 foci within 20 μm × 20 μm boxes of leptotene nuclei (Fig. 4 A and E). We then rotated the RAD51AP1 image within each box by 180° and rescored foci that colocalized with axes and DMC1 (Fig. 4F). Upon rotating the DMC1 image as a control analysis, the count decreased from 41 ± 18 axis-associated DMC1 foci in the true images to 12 ± 4 axis-associated foci in the rotated images, validating this method (Fig. 4I, P = 0.0013, Mann–Whitney U test). In contrast, the spreads with the rotated RAD51AP1 channel displayed on average 35 ± 14 axis-associated RAD51AP1 foci, which was statistically indistinguishable from the 30 ± 8 observed in the unrotated nuclei (Fig. 4I, P = 0.3696). Thus, colocalization of RAD51SP1 foci with axes may not always reflect specific association. Importantly, when we counted DMC1 and RAD51AP1 colocalized foci before and after 180° rotation of the RAD51AP1 image, the number of colocalized DMC1 decreased from 21 ± 11 to 7 ± 3 (P = 0.0027). This corresponds to a decrease from 48 ± 20% of DMC1 foci that colocalized with RAD51AP1 foci to 25 ± 15% (Fig. 4J, P = 0.0043). Thus, DMC1 foci colocalize with RAD51AP1 foci much more frequently than expected by chance.
RAD51AP1 and RAD51 colocalize to DNA damage in mitotic cells (7, 8, 16). Our analysis of 14 leptotene spermatocytes for the focus pattern of RAD51AP1 with RAD51 in meiotic chromatin spreads found frequent colocalization of RAD51AP1 with RAD51 (Fig. S6 A–D primary). Specifically, the boxed areas of these nuclei had 64 ± 17 RAD51 foci and 84 ± 18 class I RAD51AP1 foci (Fig. S6I). The fraction of RAD51 foci positive for RAD51AP1 was 49 ± 17% (Fig. S6J), a number that is very similar to the fraction of DMC1 foci positive for RAD51AP1. Importantly, upon rotation of the RAD51AP1 image (Fig. S6 I and J), the number of colocalized RAD51 foci decreased from 30 ± 9 to 7 ± 2 (P = 0.0004). RAD51AP1 foci therefore colocalized with RAD51 more frequently than expected by chance. We also established the specificity of the anti-DMC1 and anti-RAD51 antibodies by pretreating it with purified DMC1 and RAD51 proteins, respectively (Fig. S7).
Thus, RAD51AP1 foci colocalize frequently with either DMC1 or RAD51 (or conceivably both), and the RAD51AP1-DMC1 association appears to be as intimate as the RAD51AP1-RAD51 association. Because the antibodies against RAD51 and DMC1 were both raised in rabbit, we have been unable to perform double staining of RAD51-DMC1 or triple staining of RAD51-DMC1-RAD51AP1 to ascertain whether RAD51AP1 forms a tripartite complex at DSB sites.
Discussion
Published studies have provided ample evidence for a role of RAD51AP1 in mitotic HR via enhancement of the recombinase activity of RAD51 (7, 8). The molecular basis for this enhancement has not yet been determined, although it has been suggested that, through its DNA structure specific binding activity, RAD51AP1 may stabilize D loops made by RAD51 (7). We have shown here that RAD51AP1 also physically interacts with and stimulates the recombinase activity of DMC1. Using a variety of biochemical tools, we have provided evidence that RAD51AP1 synergizes with the DMC1 presynaptic filament to capture duplex DNA and assemble the synaptic complex. These attributes of RAD51AP1 are likely dependent on its duplex-DNA binding activity (8). Importantly, using naturally occurring isoforms of RAD51AP1 and mutant variants of one of these isoforms, we have furnished evidence that the functional interaction of RAD51AP1 with DMC1 is dependent on complex formation between the two HR factors and that distinct epitopes in RAD51AP1 mediate its respective interactions with DMC1 and RAD51. Interestingly, a single RAD51AP1 isoform that is most similar to human isoform 2 is expressed in the mouse testis. Importantly, meiotic chromatin spreads showed colocalization of RAD51AP1 with DMC1 in leptonema. Even though we have presented biochemical results and cytological evidence to suggest that RAD51AP1 physically and functionally interacts with DMC1, in vivo studies entailing animal models will be needed to determine if these interactions are biologically meaningful.
Despite the critical role of DMC1 in the formation of meiotic chromosome crossovers needed for the proper disjunction of homologous chromosomes in meiosis I, much remains to be learned about its functional partners (2). The HOP2-MND1 complex is one such DMC1 partner. Biochemical studies have shown that, like RAD51AP1 (7, 8), HOP2-MND1 preferentially binds dsDNA over ssDNA (11, 12) and promotes DMC1-mediated D-loop formation via functional synergy with the DMC1 presynaptic filament in duplex capture and synaptic complex assembly (12). We have found no evidence for physical interaction of RAD51AP1 and HOP2-MND1 or functional synergy of these two factors in the enhancement of the DMC1-mediated D-loop reaction. Moreover, an excess of HOP2-MND1 is unable to prevent the formation of the RAD51AP1-DMC1 complex. We note that cytological analyses done in S. cerevisiae and Arabidopsis thaliana have shown that MND1 localizes to chromatin as foci independently of Spo11-mediated DNA double-strand break formation (23–25). Curiously, in S. cerevisiae and A. thaliana, Hop2-Mnd1 does not seem to colocalize significantly with Rad51 or Dmc1, implying a transient nature of their interaction or that only a small fraction of Hop2-Mnd1 is available for the enhancement of Rad51 and Dmc1 (24, 25). In addition to functioning as a cofactor of DMC1, HOP2-MND1 could also fulfill other roles in meiotic chromosome metabolism. The precise relationship between RAD51AP1 and HOP2-MND1, for instance, whether they provide the same recombinase enhancement function but in distinct sites of the meiotic chromatin, remains to be determined.
Materials and Methods
Purification of HR Proteins.
GST- and MBP-tagged RAD51AP1 isoforms and mutants were expressed in Escherichia coli and purified as described (8). Human DMC1, SpDmc1, and ScRad54 were purified as described previously (26, 27).
Affinity Pulldown.
The indicated GST-tagged or MBP-tagged RAD51AP1 isoform (5 μg) was incubated with either (His)6-DMC1 or SpDmc1 (5 μg each) in 30 μL of buffer P (25 mM Tris-HCl, pH 7.5, 60 mM KCl, 1 mM 2-mercaptoethanol) for 30 min at 4 °C. After being mixed with 7 μL of glutathione resin (Amersham) or amylose resin (New England Biolabs) for 30 min at 4 °C to capture the tagged RAD51AP1 and associated proteins, the beads were washed three times with 30 μL of the same buffer and then treated with 20 μL of 2% SDS to elute proteins. The supernatant, last wash, and SDS eluate, 10 μL each, were analyzed by SDS-PAGE.
D-Loop Assay.
The D-loop assay was conducted at 37 °C, as described (22, 25). Briefly, the 32P-labeled 90-mer oligonucleotide substrate (2.4-μM nucleotides) was incubated for 5 min with DMC1 or SpDmc1 (0.8 μM) in 10.5 μL of buffer D (35 mM Tris-HCl, pH 7.5, 50 mM KCl, 2 mM ATP, 4 mM MgCl2, 100 μg/mL BSA, and 1 mM dithiothreitol), followed by the addition of the indicated amount of RAD51AP1 or ScRad54 in 1 μL and a 5-min incubation. pBluescript replicative form I DNA (35-μM base pairs) was then incorporated in 1 μL, and after a 20-min incubation, the reaction mixtures were analyzed (8, 11).
Duplex Capture Assay.
The assay followed our published procedure (11). To assemble the presynaptic filament, 4 μL of magnetic beads containing 5′-biotinylated 83-mer dT ssDNA (60 ng, corresponding to 9 μM nucleotides) were incubated with 2.7 μM of DMC1 or SpDmc1 in 18 μL of buffer D for 5 min at 37 °C. The beads were captured with the Magnetic Particle Separator, washed once with 20 μL of buffer D, and then resuspended in 15 μL of the same buffer. Following the incorporation of the indicated amount of RAD51AP1 and a 5-min incubation at 37 °C, the beads were again captured, washed once with 150 μL of buffer D, and resuspended in 18 μL of buffer D. The reaction was completed by adding radiolabeled dsDNA (4-μM base pairs) with or without radiolabeled ssDNA (4-μM nucleotides) in 2 μL. The reaction mixtures (20 μL final volume) were incubated for 10 min at 37 °C with gentle mixing every 30 s. The beads were captured, washed twice with 200 μL of buffer D, and the bound proteins and radiolabeled DNA were eluted with 20 μL of 2% SDS. The supernatant and SDS eluate were resolved in a 10% polyacrylamide gel in TBE buffer (90 mM Tris-HCl, 70 mM boric acid, pH 8.0, 2 mM EDTA), and the DNA species were visualized and quantified by phosphorimaging analysis.
Assay for Synaptic Complex Formation.
To assemble the presynaptic filament, the 60-mer oligonucleotide (12) (12-μM nucleotides) were incubated with 4 μM of DMC1 or SpDmc1 in 8 μL of buffer D containing 2 mM ATP for 5 min at 37 °C. The indicated amount of RAD51AP1 isoform or mutant was added in 1 μL, and, after another 5-min incubation at 37 °C, EcoR1-linearized pUC19 DNA (corresponding to 83 μM of nucleotides) was added in 1 μL, followed by the incorporation of 6.5 units of Ssp1 and a further incubation for 10 min at 37 °C. The reaction mixtures were deproteinized with SDS and proteinase K (0.1% and 0.5 mg/mL, respectively) for 5 min at 37 °C before being resolved in a 0.9% agarose gel in TAE buffer (40 mM Tris, 20 mM NaOAc, pH 7.5, 2 mM EDTA). The DNA species were stained with ethidium bromide and recorded in a BioRad gel documentation station.
Anti-RAD51AP1 Antibodies.
MBP-tagged mouse RAD51AP1 was expressed in and purified from E. coli by following the procedures employed for MBP-tagged human RAD51AP1 (8). Following cleavage of the MBP tag, the mouse RAD51AP1 protein was used to generate guinea pig antiserum at Cocalico Biologicals, Inc. This GP49 antiserum was affinity-purified using cyanogen bromide activated Sepharose 4B (Pharmacia-LKB) beads covalently conjugated to the mouse MBP-RAD51AP1 protein, as previously described (27).
Immunofluorescence.
Spermatocytes were collected from juvenile (15 dpp) mouse testes and prepared for surface spreading as previously described (28). Slides were blocked in antibody dilution buffer (1×PBS, 2 mg/mL BSA, 0.05% Tween-20, 0.2% gelatin) for 10 min at 25 °C, and then incubated overnight at 4 °C with 1∶200 guinea pig anti-RAD51AP1 antibody (GP49), 1∶100 rabbit anti-DMC1 antibody (H-100, Santa Cruz), or 1∶650 mouse anti-SYCP3 antibody (D-1, Santa Cruz). Primary antibodies were detected using TexasRed goat anti-mouse IgG, Cy5 goat anti-rabbit IgG, or AlexaFluor-488 goat anti-guinea pig IgG (Invitrogen). Slides were mounted in Vectashield mounting medium (Vector) with 50 μg/mL DAPI. Images were captured using an Axio2 microscope (Zeiss) connected to a CCD camera and processed using the SlideBook software package (Intelligent Imaging Innovations). Leptonema was defined as having short, unsynapsed stretches of SYCP3. Early zygonema was defined as having some (< 30%) synapsed SYCP3 stretches. Only DMC1 foci that colocalized with SYCP3 staining were counted. The number of colocalizing AP1-DMC1 and AP1-RAD51 foci was scored without restriction to the subset of foci that share the same centroid. Fluorescence channels were aligned by imaging 0.2-μm Tetraspeck beads (Invitrogen). Images were quantified using the Align Channel function in the Slidebook software.
Statistical Analysis of Foci Colocalization.
The significance of DMC1 and RAD51AP1 colocalization was tested by performing the Mann–Whitney U test on the percentage of colocalized foci in the real image versus the rotated image (after RAD51AP1 channel was rotated 180°). This test, appropriate to small samples, was chosen as an alternative of the Student test because the two datasets are nonparametric. The DMC1 dataset was drawn from a total of eight spermatocytes (N = 8), and the RAD51 dataset was drawn from a total of nine spermatocytes (N = 9). The Prism 5 software was used to generate two-tailed P values.
Supplementary Material
Acknowledgments.
SpDmc1 was a kind gift from Peter Chi (Department of Molecular Biophysics and Biochemistry, Yale University School of Medicine, New Haven, CT). We thank Jim Dowdle for helping to prepare immunofluorescence slides. This work was supported by National Institutes of Health Grants R01ES015252, R01HD040916, R01ES07061, P01CA092584, and R01-CA120315 and Susan G. Komen for the Cure Foundation postdoctoral fellowship PDF0706844.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016454108/-/DCSupplemental.
References
- 1.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi M, DasGupta C, Radding CM. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell. 1983;34:931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- 5.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 6.Kovalenko OV, Golub EI, Bray-Ward P, Ward DC, Radding CM. A novel nucleic acid-binding protein that interacts with human rad51 recombinase. Nucleic Acids Res. 1997;25:4946–4953. doi: 10.1093/nar/25.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modesti M, et al. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Wiese C, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riddles PW, Lehman IR. The formation of paranemic and plectonemic joints between DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J Biol Chem. 1985;260:165–169. [PubMed] [Google Scholar]
- 11.Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–1757. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–1766. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrin LJ, Camerini-Otero RD. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- 14.Kovalenko OV, Wiese C, Schild D. RAD51AP2, a novel vertebrate- and meiotic-specific protein, shares a conserved RAD51-interacting C-terminal domain with RAD51AP1/PIR51. Nucleic Acids Res. 2006;34:5081–5092. doi: 10.1093/nar/gkl665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetz P, Chandley AC, Speed RM. Morphological and temporal sequence of meiotic prophase development at puberty in the male mouse. J Cell Sci. 1984;65:249–263. doi: 10.1242/jcs.65.1.249. [DOI] [PubMed] [Google Scholar]
- 16.Mizuta R, et al. RAB22 and RAB163/mouse BRCA2: Proteins that specifically interact with the RAD51 protein. Proc Natl Acad Sci USA. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarsounas M, Morita T, Pearlman RE, Moens PB. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol. 1999;147:207–220. doi: 10.1083/jcb.147.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: Occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107:2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- 19.Lammers JH, et al. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moens PB, Pearlman RE. Chromatin organization at meiosis. Bioessays. 1988;9:151–153. doi: 10.1002/bies.950090503. [DOI] [PubMed] [Google Scholar]
- 21.Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kee K, Protacio RU, Arora C, Keeney S. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J. 2004;23:1815–1824. doi: 10.1038/sj.emboj.7600184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 24.Zierhut C, Berlinger M, Rupp C, Shinohara A, Klein F. Mnd1 is required for meiotic interhomolog repair. Curr Biol. 2004;14:752–762. doi: 10.1016/j.cub.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 25.Vignard J, et al. The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 2007;3:1894–906. doi: 10.1371/journal.pgen.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi P, et al. Functional interactions of meiotic recombination factors Rdh54 and Dmc1. DNA Repair. 2009;8:279–284. doi: 10.1016/j.dnarep.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigurdsson S, Van Komen S, Petukhova G, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2002;277:42790–42794. [Google Scholar]
- 28.Barchi M, et al. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol. 2005;25:7203–7215. doi: 10.1128/MCB.25.16.7203-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.