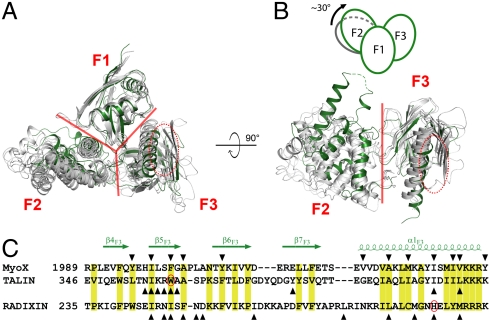
Fig. 4.
Comparison of the structure of MyoX FERM with those of other FERM domains. (A and B) MyoX FERM and other FERM domains, including radixin (PDB ID code 1GC6), merlin (1H4R), protein 4.1 (2HE7), and FAK (2AEH) are overlapped by superimposing their F1 lobes. The borders between each pair of the three lobes are indicated with red lines. The MyoX FERM domain is colored in green while other FERM domains are in gray. The αβ-groove in the F3 lobe is indicated by red ovals. In B, the superimposed F1 lobes are omitted for better appreciation of the conformational differences of the F2 lobes in different FERM domains. The schematic diagram shows the approximately 30° rotation of the MyoX F2 lobe. (C) Structural based sequence alignment of the αβ-grooves from MyoX, talin, and radixin. The conserved residues are labeled with yellow boxes. The residues of which side chains are involved in their respective target interactions are marked by triangles. The two residues highlighted in Fig. 5 C and D are also circled.