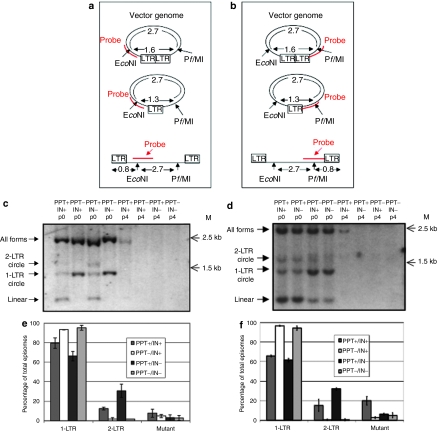
Figure 3.
Analyzing the effect of the polypurine tract (PPT) deletion on lentiviral vector episome formation. (a,b) Outline of Southern-blot analysis used to characterize vector provirus and episomes by probing at the (a) 5′ and (b) 3′ regions of the genome. After extracting vector DNA from transduced cells, digestion with EcoNI and PflMI, and probing with either (a) a KasI/BamHI fragment of at PPT+ vector (vTK945) or (b) a AfeI/EcoRV fragment of vTK945, bands indicative of total episomes and integrated provirus (2.7 kb), 2-long terminal repeat (LTR) circles (1.6 kb), 1-LTR circles (1.3 kb), and linear episomes (0.8 kb) were determined. (c) Cells were transduced with a PPT+ vector (vTK945), integrating (lanes 1 and 5) or nonintegrating (lanes 3 and 7), or with a PPT− vector (vTK1179), integrating (lanes 2 and 6) or nonintegrating (lanes 4 and 8) and total DNA was extracted from transduced cells 3 days (no passages) (lanes 1–4) or ~14 days (four passages (p4)) (lanes 5–8) post-transduction, digested as shown in a and b, and analyzed by Southern blot, using the EcoNI-spanning probe complementary to the 5′ region of the virus. (d) Cells were transduced with the PPT+ vector, integrating (lanes 1 and 5) or nonintegrating (lanes 2 and 6), or with the PPT− vector, integrating (lanes 3 and 7) or nonintegrating (lanes 4 and 8) and total DNA was extracted from transduced cells 3 days (no passages) (lanes 1–4) or ~14 days (p4) (lanes 5–8) post-transduction, digested as in (a,b), and analyzed by Southern blot, using the PflMI-spanning probe complementary to the 3′ region of the virus. For c,d, bands corresponding to total vector genomes, 2-LTR circular, 1-LTR circular, and linear episomal genomes, as outlined in a,b, respectively, are shown. (e) Sixteen hours after transduction with a non-self-inactivating (non-SIN), PPT-deleted shuttle vector (vTK1074) and its PPT-positive equivalent (vTK459), which were packaged with both functional integrase (IN+) and mutant integrase (IN−), episomes were harvested from 293T cells and analyzed by shuttle–vector assay; each transduction was performed in triplicate. (f) Episomes were harvested from 293T cells 16 hours after transduction with a SIN, PPT-deleted shuttle vector (vTK1046) and its PPT-positive equivalent (vTK1054), which were packaged with IN+ and IN−, and analyzed by shuttle–vector assay; each transduction was performed in triplicate. For e,f, the data are presented as percentages of 1-LTR, 2-LTR, and mutant forms out of the number of total episomes.