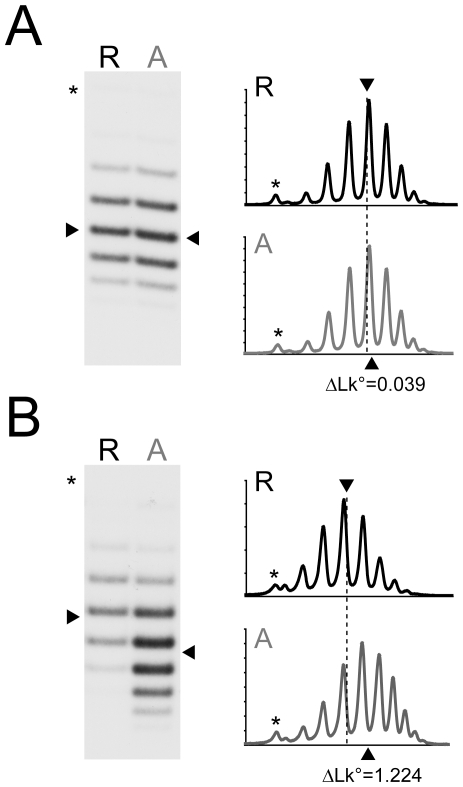
Figure 4. Effect of UASp mutations on PHO5 promoter nucleosome loss.
Nucleosome loss at the PHO5 promoter was measured by topology analysis [29], in UASp1 and UASp2 deletion mutants. Gene circles were formed in vivo by site-specific recombination of the PHO5 locus [29]. Gene circle topoisomers were fractionated by chloroquine gel electrophoresis, blotted and hybridized with a 32P-labeled DNA probe spanning the PHO5 gene. (A) Topoisomer distributions of PHO80 (R) and pho80Δ (A) PHO5 gene circles isolated from UASp1Δ cells. Pho80 is an inhibitor of the PHO signaling pathway. In its absence Pho4 is constitutively nuclear. (B) Topoisomer distributions of PHO80 (R) and pho80Δ (A) PHO5 gene circles isolated from UASp2Δ cells. The intensity profiles of distributions are shown on the right next to the autoradiographs. Distribution centers are indicated by arrowheads. The center of the topoisomer distribution for the repressed gene is additionally marked by a dashed line in the intensity profiles. Numbers indicate the linking difference, ΔLk°, between distribution means (arrowheads), which corresponds to the average number of nucleosomes lost from the activated promoter [29]. Wild type PHO5 gene circles have a linking difference of 1.85 [8], [29]. Positions of nicked circles are indicated by (*).