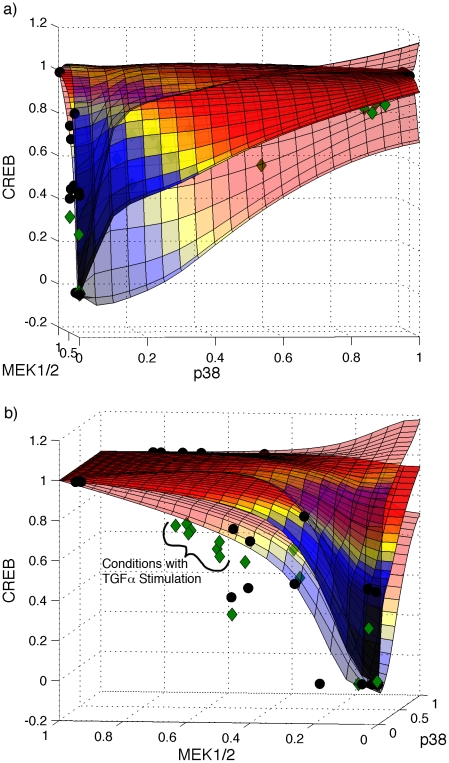
Figure 7. Transfer functions predicted by trained cFL models.
The output value of the CREB node was predicted by computationally simulating each individual model in the family of cFL models with 441 combinations of p38 and MEK1/2. Three-dimensional plots were generated in MATLAB showing the average prediction (opaque surface) as well as the average prediction plus or minus the standard deviation of the predicted value (semi-transparent surfaces). The training data (black circles) and validation data (green diamonds) are also plotted. The 3-D plots have been rotated to highlight the influence of either (a) p38 or (b) MEK1/2. The predicted transfer functions agree with the validation data reasonably well except for the overestimation of CREB activation for conditions with TGFα stimulation as one of the ligands.