Figure. 6.
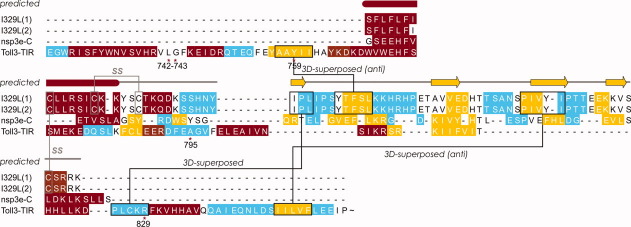
Sequence alignment by secondary-structure match of the two best models of pI329L with the SARS-CoV domain (nsp3e-C) and TLR3-TIR. Relevant TLR3 key residues are marked (*) and numbered according to Ref.4; the last ∼50 residues of this sequence have been omitted for simplicity. The two disulfide (SS) bridges were assigned on proximity criteria. A three-dimensional (3D)-superposition line indicates that two regions that are distant in terms of sequence alignment fall on top of each-other when the 3D-structures are superposed. An “anti” superposition means that two β-strands superpose head-to-tail. The I-TASSER predicted secondary structure for pI329L runs on top of the alignment: rod, helix; arrow, strand; and thin line, coil. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]