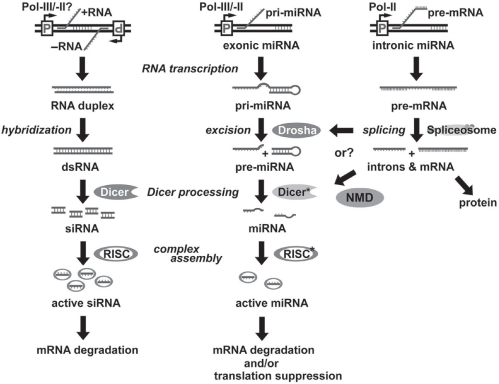
Figure 2.
Comparison of various biogenesis and PTGS mechanisms among siRNA, intergenic (exonic) miRNA and intronic miRNA. SiRNA is formed by annealing of two perfectly complementary short RNAs transcribed from two opposite promoters (remains to be determined), and then further processed into 19–22 bp double-stranded duplexes by RNaseIII-familial endonucleases, Dicer. Intergenic (exonic) miRNA, eg, lin-4 and let-7, is directly transcribed as a long single-strand noncoding RNA precursor (pri-miRNA), probably by either a Pol-II or a Pol-III RNA promoter, whereas intronic miRNA is mainly transcribed by the Pol-II promoter of its encoded gene and co-existed within the intron region of the encoded gene transcript (pre-mRNA). After pre-mRNA splicing, the spliced intron functions as a pri-miRNA to form intronic miRNA. In the nucleus, the pri-miRNA is excised by either a Drosha RNase (intergenic miRNA) or spliceosomal/NMD components (intronic miRNA) to form hairpin-like miRNA precursors (pre-miRNA) and then exported to cytoplasm for further processing by Dicer* to form mature miRNAs. The Dicers for siRNA and miRNA pathways may be different. For instance, some spliceosomal/NMD components are likely involved in the maturation of intronic miRNA but not siRNA. All three small regulatory RNAs are finally incorporated into a RNA-induced silencing complex (RISC), which contains either strand of siRNA or the single-strand of miRNA. The effect of miRNA is considered to be more specific and less adverse than that of siRNA because only one strand is involved. On the other hand, siRNAs primarily trigger mRNA degradation, whereas miRNAs can induce either mRNA degradation or translational suppression, depending on the sequence complementarity to their targeted gene transcripts.