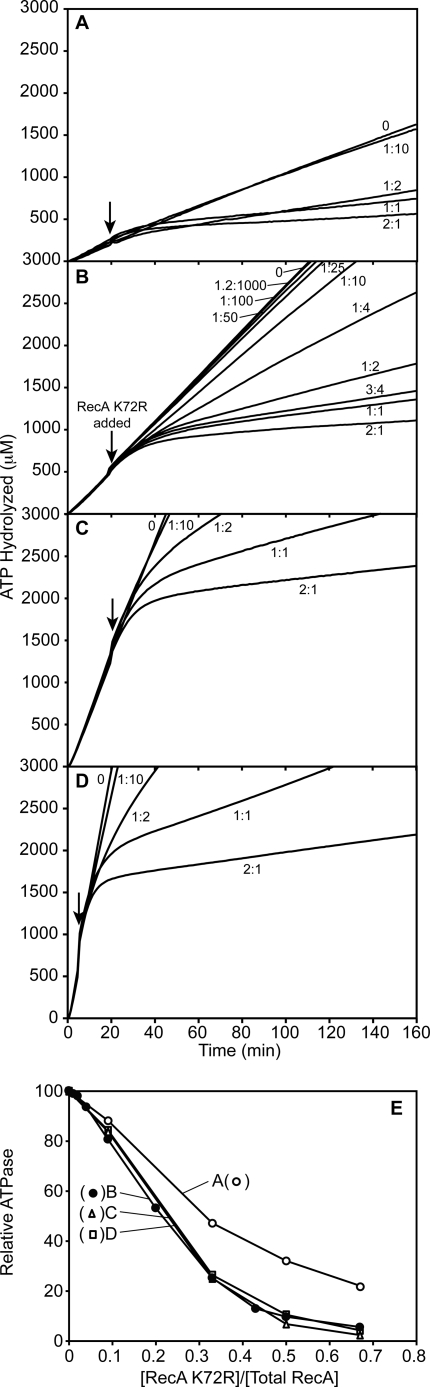
FIGURE 6.
A significant proportion of RecA K72R is needed to fully inhibit ATP hydrolysis by established wild type RecA filaments on cssDNA. Wild type RecA filaments formed on cssDNA were challenged with varying concentrations of RecA K72R, and the resultant decrease in the rate of wild type RecA-catalyzed ATP hydrolysis was monitored over time. ATPase assays were carried out as described under “Experimental Procedures.” The reactions all contained 3 mm ATP. Other conditions for panels A, B, C, and D, respectively, were: M13mp18 cssDNA, 2, 4, 8, and 16 μm; wild type RecA, 0.67, 1.33, 2.66, and 5.32 μm; SSB, 0.2, 0.4, 0.8, and 1.6 μm. In each experiment the RecA K72R was varied in concentration from 0 μm to twice the concentration of wild type RecA. RecA K72R concentrations are presented as a ratio of RecA K72R to wild type RecA. Wild type RecA filaments were allowed to hydrolyze ATP for 20 min before RecA K72R addition in panels A, B, and C and 5 min in panel D. In panel E, the wild type RecA ATPase in the unchallenged reaction is taken as 100%, and the proportional decline in the ATPase rate in the new steady state is plotted as a function of the mutant to wild type protein ratio after the addition of RecA K72R. The new steady-state rates were defined beginning 30 min after the mutant protein challenge in panels A and B and beginning at 20 min for most reactions (1:2, 1:1, and 2:1) in panels C and D. In the latter two panels, the new rate after a 1:10 challenge was determined during the 10 min immediately before the exhaustion of the ATP regeneration system.