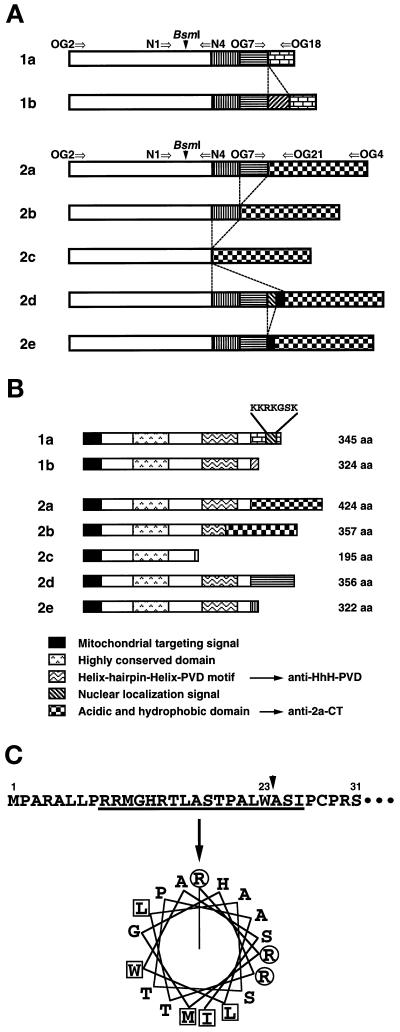
Figure 1.
Structures of multiple human OGG1 mRNAs and their products. (A) Alternatively spliced forms of OGG1 mRNAs. Each box represents a region alternatively joined in each OGG1 mRNA. The open box represents the sequence corresponding to exons 1 to 3 of the human OGG1 gene (Aburatani et al., 1997), in which alternative splicing was not observed. Arrows indicate primers used for RT-PCR analysis. These sequence data of human OGG1 cDNA are available from DNA Data Bank of Japan under the following accession numbers: 1b, AB019532; 2b, AB019528; 2c, AB019529; 2d, AB019530; and 2e, AB019531. (B) Structural features of different human OGG1 proteins. All forms of the OGG1 proteins contain a putative MTS at the common N-terminal region. There is a NLS only in the C-terminal end of OGG1–1a. The region exhibiting the highest homology to the corresponding region of yeast Ogg1 protein (aa 128–156) is shown as the highly conserved region. Anti–HhH-PVDrecognizes the region containing HhH-PVD motif (aa 221–290), and anti–2a-CT recognizes the C-terminal regions of both OGG1–2a (aa 322–424) and -2b (aa 255–357). (C) Putative mitochondrial targeting signal for OGG1 proteins. Using the MitoPlot II program, we predicted that the residues from 9 to 26 at the common N-terminal region of OGG1 proteins form an amphipathic helix (angle for the helical wheel is 95°), with three arginine residues but no acidic residue and values of 6.57 for hydrophobic moment (μH), 4.33 for maximum hydrophobicity (Hmax). The MTS may be processed at residue 23 (W) after being translocated into mitochondria (arrow). Basic residues (R) are shown in circles, and hydrophobic residues are in boxes.