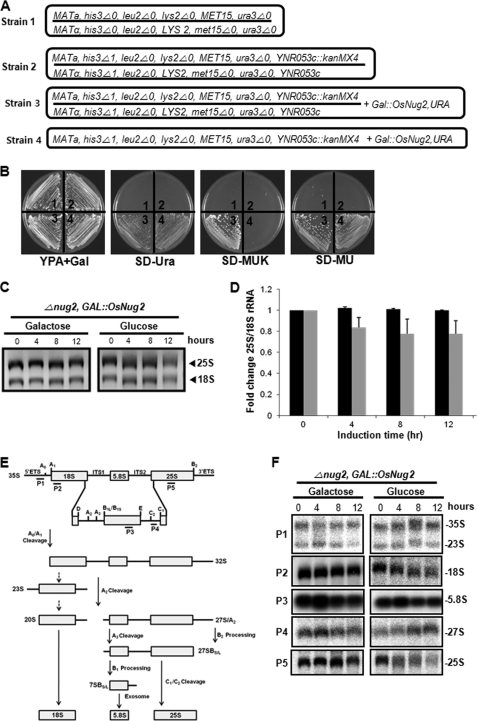
FIGURE 3.
OsNug2 complements the yeast nug2 null mutation. A, the strains used are as follows: strain 1, S. cerevisiae BY4743; strain 2, S. cerevisiae Y26080; strain 3, S. cerevisiae Y26080 with Gal::OsNug2; strain 4, S. cerevisiae Y26080 nug2 null mutant with Gal::OsNug2 from strain 3. B, the growth phenotype of strains 1–4 (numbered at the center of each sector in the Petri dish) were cultivated on different media. All media includes galactose. C, total RNAs were extracted at different time intervals from strain 4 cultures under OsNug2-inducible conditions on galactose-containing medium, and under OsNug2-suppression conditions on glucose-containing medium. Samples were separated on a 1.5% agarose-formaldehyde gel and stained with ethidium bromide. An equal amount (1 μg) of total RNA was loaded in each lane. D, the 25S/18S rRNA ratios are dependent upon induction time. The graph represents fold-changes in the 25S/18S rRNA ratio of Strain 4 cultured as described above for C, with OsNug2-inducible conditions (black) and OsNug2-suppression conditions (gray). These data were obtained from three independent experiments. E, schematic diagram showing the pre-rRNA and rRNA processing pathways in S. cerevisiae, and indicating the positions of hybridization probes. F, different RNAs were extracted from strain 4 cultured as described above in C and hybridized using gene-specific oligonucleotide probes. The probes used for RNA gel blot analysis are shown in E as P1 to P5 and were prepared as described previously (55). An equal amount (1 μg) of total RNA was loaded in each lane.