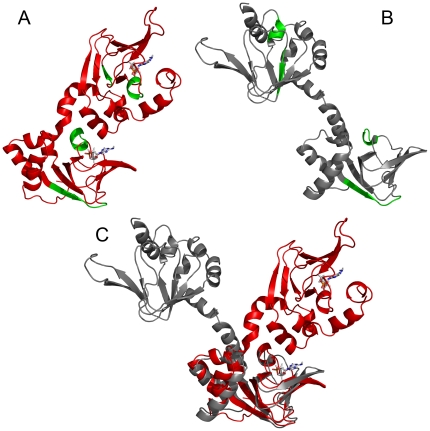
Figure 6. Aggregation prone areas as indicated by TANGO and high temperature MD.
A) Structural mapping of the β-type aggregation motifs implicated by the TANGO algorithm onto the structure of bovine RIα(113–376) (PDB 1RSG) [7], regions 153–159, 201–206, 290–294 and 325–329 are shown in green. 201–206 and 325–329 constitute the B' helices of CNB domains A and B, i.e. αB':A and αB':B, respectively. B) Structure after the MD simulation at 450 K (cAMP was deleted from the complex (PDB 1RSG) at the beginning of the MD simulation; see text for details). (C) Superposition of the initial cAMP-free structure (red) and after high-temperature MD simulation (gray). The superposition was carried out by best-fitting of CNB domain A. RMSD values (C-alpha atoms) are 3.9 Å for the CNB A domains and 15.7 Å for the entire protein. Note that the change in relative orientation of the domains around the interdomain C/C' helices occurs without any significant changes in the structure of the domains β-sandwich.