Abstract
A system was devised that enables quantitative, ligand-dependent exponential amplification for various ligands that can be recognized by an RNA aptamer. The aptamer is linked to an RNA enzyme that catalyzes the joining of two oligonucleotide substrates. The product of this reaction is another RNA enzyme that undergoes self-sustained replication at constant temperature, increasing in copy number exponentially. The concentration of the ligand determines the amount of time required for the replication products to reach a threshold concentration. A standardized plot of time to threshold versus ligand concentration can be used to determine the concentration of ligand in an unknown sample. This system is analogous to quantitative PCR, linking rare recognition events to subsequent exponential amplification, but unlike PCR can be applied to the quantitative detection of non-nucleic-acid ligands.
Introduction
The exponential amplification of nucleic acids has become a core technology in medical diagnostics and many routine laboratory procedures. The most widely used amplification technique is the polymerase chain reaction (PCR), together with its variants such as reverse transcription PCR, immuno-PCR, and proximity ligation, which can be used to detect and quantify target nucleic acids and proteins.1–4 A less commonly practiced amplification technique is the ligase chain reaction (LCR), which utilizes a thermostable DNA ligase and two pairs of DNA substrates.5,6 The first pair of substrates bind to the target nucleic acid and are ligated to form a complementary product; the second pair of substrates then bind to the complementary product and are ligated to form a new copy of the target nucleic acid. Both PCR and LCR require temperature cycling to bring about successive rounds of amplification. There also are RNA amplification procedures that operate at constant temperature, combining reverse transcription and forward transcription to achieve exponential amplification.7
Recently a new method for isothermal, exponential amplification of RNA was described that is analogous to LCR, but does not require any protein enzymes.8 Instead, the replicating molecules bring about their own amplification through their catalytic activity as template-dependent RNA ligases. Two RNA enzymes operate as a cross-replicating pair, each ligating two RNA substrates to form new copies of its partner. Cross-replication can be made dependent on recognition of a target ligand by linking the catalytic domain of the ligase to an aptamer domain that binds the ligand.9 This results in an “autocatalytic aptazyme” that is active only when the ligand is bound. The exponential growth rate of these molecules reflects the concentration of the ligand relative to the Kd of the aptamer-ligand interaction. The ligand can be any small molecule or macromolecule that is recognized by a corresponding RNA aptamer, thus providing a general method for the quantitative detection of target ligands.
One of the chief limitations of the autocatalytic aptazymes is that the ligand-sensing molecule must also function as a substrate for the ligase enzyme. The Km of the enzyme-substrate interaction is in the micromolar range, which may exceed the desired Kd of the aptamer-ligand interaction, the latter being determined by the concentration of ligand in the relevant analytical context. If the concentration of the ligand-sensing molecule is reduced below the Km in order to sense low concentrations of ligand, then the efficiency of amplification will be compromised. This problem is surmounted in quantitative PCR (qPCR), which employs target-sensing molecules (oligodeoxynucleotide primers) at concentrations in the micromolar range, yet can determine concentrations of a target nucleic acid in the sub-picomolar range.2,10 When carrying out qPCR, one determines the number of temperature cycles needed to reach some threshold yield of amplification products. A similar approach can be applied to the autocatalytic aptazymes, except that amplification is continuous, and therefore one must determine the amount of time required to reach the threshold.
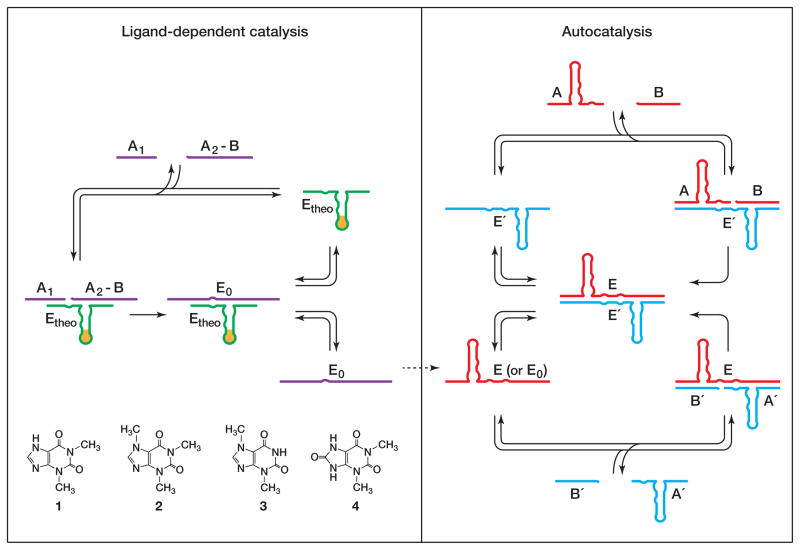
A non-replicating, ligand-dependent ligase enzyme is employed to generate a seed concentration of replicating ligase enzymes, which subsequently initiate ligand-independent exponential amplification (Figure 1). Generation of the seed is analogous to the first round of qPCR, which involves primer extension on a target nucleic acid template. Subsequent rounds of qPCR proceed in a target-independent manner, with primer extension occurring on templates that were produced during the previous rounds. With both methods, the concentration of target molecules in the reaction mixture determines the concentration of seed that is generated, which in turn determines the number of rounds of amplification needed to reach the threshold.
Figure 1.
Scheme for ligand-dependent exponential amplification, coupling ligand-dependent catalysis (left) to ligand-independent autocatalysis (right). The ligand is recognized by an aptamer domain that is linked to an RNA ligase enzyme (Etheo). In the presence of the cognate ligand 1, but not closely-related compounds 2–4, Etheo catalyzes the joining of two oligonucleotide substrates (A1 and A2-B) to form ligase enzymes (E0) that can seed exponential amplification. Exponential amplification involves two ligase enzymes (E and E′) that catalyze each other’s synthesis from a total of four component substrates (A and B, and A′ and B′, respectively).
The approach described here for the quantitative exponential amplification of nucleic acids can be extended to a broad range of targets for which a suitable aptamer can be developed. Over the past 20 years the technology for developing RNA aptamers has become increasingly routine,11 now comparing favorably with methods for generating target-specific antibodies. The present study employed the previously-described theophylline aptamer,12 which was installed adjacent to the catalytic domain of the R3C RNA ligase enzyme.13 This construct enables the quantitative detection of theophylline (1) in the concentration range of 2–500 μM. The lowest concentration of theophylline that can reliably be detected is 80-fold lower than the Kd of the aptamer-ligand interaction.
Results
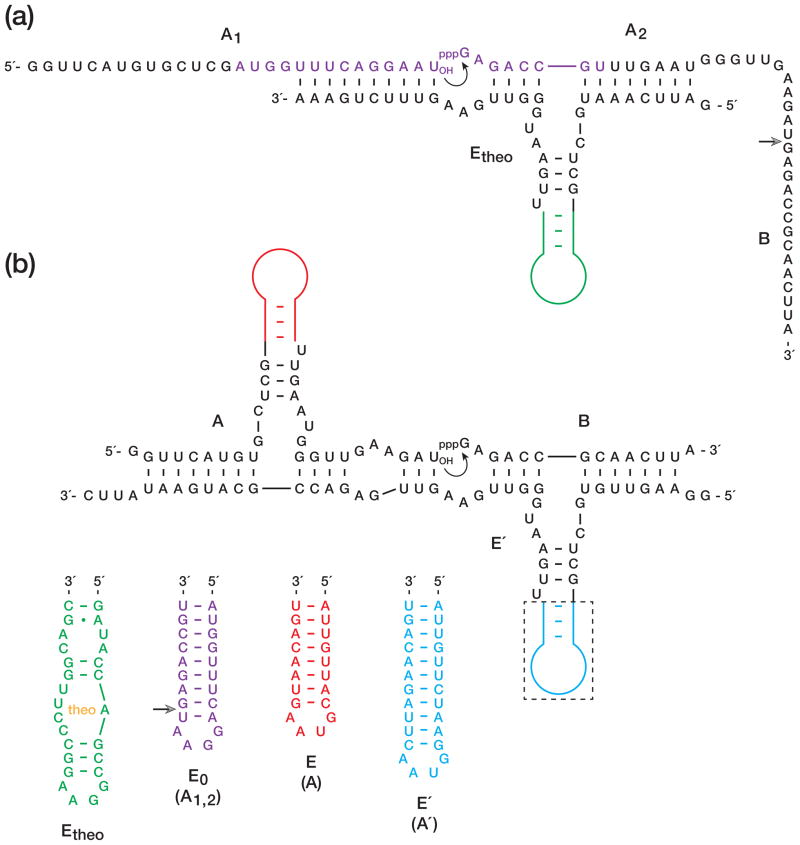
The R3C RNA ligase was converted to an aptazyme (Etheo) by replacing the central stem-loop of the enzyme by an aptamer that is specific for theophylline (Figure 2a). In the presence but not the absence of theophylline, the aptamer domain assumes a well-defined structure that supports the catalytic domain of the enzyme. The enzyme catalyzes the joining of two RNA substrates, one bearing a 3′-hydroxyl (A1) and the other bearing a 5′-triphosphate (A2-B), resulting in formation of a 3′,5′-phosphodiester and the release of inorganic pyrophosphate. The ligated product contains both portions (A1,2 and B) of an RNA enzyme (E0) that can initiate cross-replication. The site of ligation occurs within the central stem-loop of the A portion of E0, and covalent joining at this position is required for the catalytic activity of E0.
Figure 2.
Sequence and secondary structure of RNA enzymes and substrates used in this study. (a) Etheo contains an aptamer domain that binds theophylline, thereby triggering ligation of A1 and A2-B to form E0. Curved arrow indicates the site of ligation; open arrowhead indicates the junction between A2 and B. E0, in turn, catalyzes ligation of A′ and B′ to form E′. (b) E′ catalyzes ligation of A and B to form E, which behaves similarly as E0. The aptamer domain within Etheo (green) is replaced by simple hairpins in E0 (purple), E (red), and E′ (blue). Open arrowhead indicates the junction between A1 and A2.
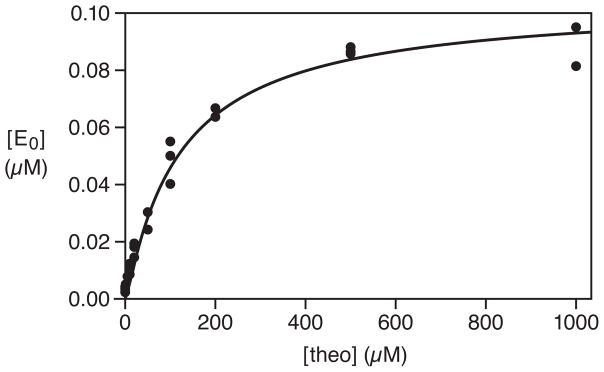
The theophylline-dependent ligation reaction employed 5 μM Etheo, 0.5 μM each of A1 and A2-B, and 25 mM MgCl2, and was incubated at pH 8.5 and 42 °C. Under these conditions, the Kd for binding of Etheo and theophylline is 160 μM (Supplementary Figure 1). Various concentrations of theophylline were tested, focusing especially on the range of 2–500 μM. The yield of E0 was determined after 20 min incubation, which in the presence of saturating theophylline resulted in consumption of ~20% of the substrates. The yield as a function of theophylline concentration fit well to a saturation plot (r = 0.993), with an apparent Kd of 130 μM and maximum extent of 0.092 μM (Figure 3). The yield of E0 after 20 min was normalized to the maximum extent to obtain the fractional saturation.
Figure 3.
Saturation plot of the ligand-dependent reaction catalyzed by Etheo. The yield of E0 was determined after 20 min in the presence of various concentrations of theophylline. These data were fit to the equation: , where ε is the yield in the absence of theophylline (0.004 μM) and [E0]max is the yield at saturation (0.092 μM).
The products of theophylline-dependent RNA ligation then were used to initiate cross-replication. Cross-replication employs two RNA enzymes (E and E′) that catalyze each other’s synthesis from a total of four component substrates (A′ + B′ → E′ and A + B → E, respectively). E0 behaves similarly to E in that it too catalyzes the joining of A′ and B′ to form E′ (Figure 2b). At the outset there are no copies of E or E′ in the reaction mixture, but both are generated exponentially as cross-replication proceeds. The amplification profile for E (and similarly for E′) can be described by the equation: , where a is the final extent, b is the degree of sigmoidicity, and c is the exponential growth rate.
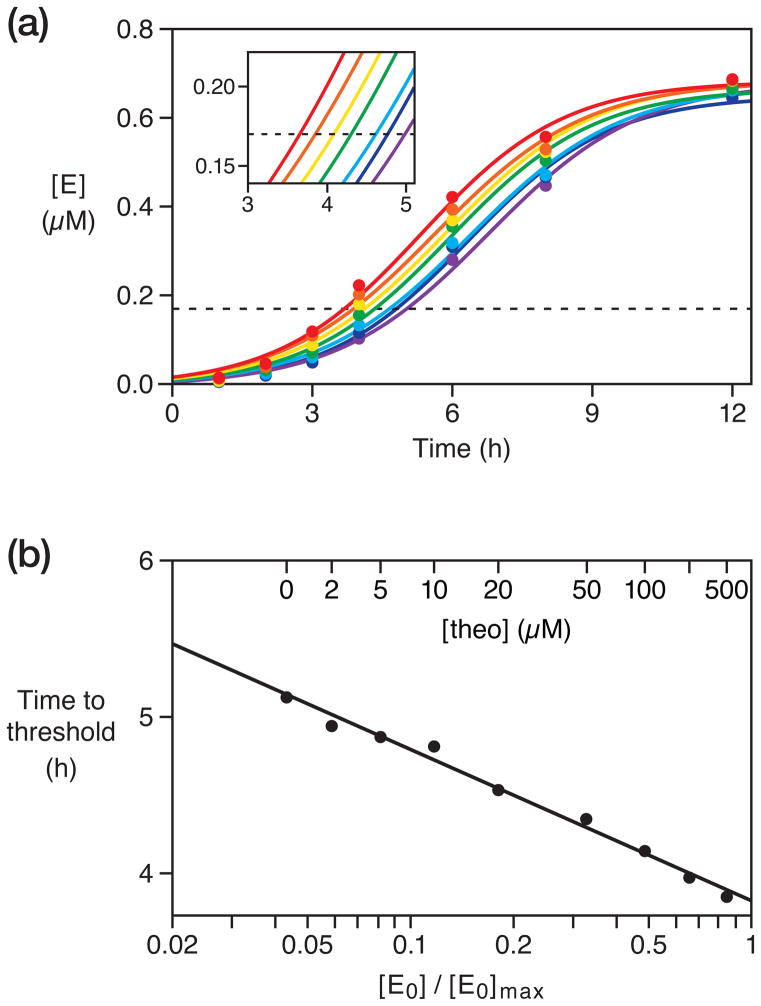
The amplification mixture contained the products of theophylline-dependent ligation, 1 μM each of A and B, 1.5 μM each of A′ and B′, and 25 mM MgCl2, and was incubated at pH 8.5 and 42 °C. With increasing concentrations of theophylline, which resulted in increasing concentrations of E0, the production of E and E′ occurred more rapidly (Figure 4a). The final extent of amplification of E was 0.68 μM, limited by consumption of the A and B substrates. A threshold concentration of 0.17 μM was chosen, corresponding to 25% of the final extent, and the amount of time required to reach this threshold was calculated for each amplification profile (Figure 4a, inset). A semi-log plot of time to threshold versus fractional saturation was linear (r = 0.995) over a saturation range of 0.06–0.84, corresponding to 2–500 μM theophylline (Figure 4b). Amplification occurs even in the absence of theophylline, with a time to threshold indicative of an apparent saturation value of 0.04. As in qPCR, target-independent initiation of exponential amplification sets a lower bound for the dynamic range of the assay. The upper bound is determined by the maximum concentration of aptazyme-ligand complexes, analogous to the maximum concentration of primer-target complexes in qPCR.
Figure 4.
Quantitative, isothermal, ligand-dependent exponential amplification. (a) The yield of E (and E′) was monitored over time in the presence of 0 (purple), 2 (blue), 10 (cyan), 20 (green), 50 (yellow), 100 (orange), or 200 (red) μM theophylline. These data were fit to the equation: , where a is the final extent, b is the degree of sigmoidicity, and c is the exponential growth rate. A threshold (dashed line) was set at 0.17 μM E, corresponding to 25% of the maximum extent of amplification. Inset shows each amplification profile as it crossed the threshold. Amplification profiles for 5 and 500 μM theophylline are omitted for clarity. (b) Time to threshold as a function of the concentration of E0, which reflects the concentration of theophylline. Linear regression coefficient was 0.995.
The system for coupled ligand-dependent catalysis and exponential amplification also was used to measure the concentration of theophylline in 10% human serum that had been deproteinized to remove nucleases. The components of serum were slightly inhibitory to RNA catalysis, which required increasing the time for ligand-dependent formation of E0 from 20 to 30 min to achieve comparable yields. The yield of E0 as a function of theophylline concentration again fit well to a saturation plot, with an apparent Kd of 170 μM and maximum extent of 0.12 μM (data not shown). Subsequent exponential amplification was performed as described above, and the time to threshold was determined for various concentrations of theophylline (Supplementary Figure 2a). A semi-log plot of time to threshold versus fractional saturation again was linear (r = 0.997) over a theophylline concentration range of 2–500 μM (Supplementary Figure 2b).
Efforts were made to understand the basis for exponential amplification that occurs even in the absence of theophylline. Etheo is strongly dependent on theophylline for its ability to ligate A1 and A2-B to form E0 (Figure 3), but it is possible that Etheo also can ligate A and B to form E directly, perhaps in a theophylline-independent manner. This was found not to be the case. The addition of 5 μM Etheo (but no E0) to an amplification mixture that contained either 0 or 1000 μM theophylline had no influence on the exponential amplification profile (Supplementary Figure 3).
Alternatively, it is possible that the unligated components of E0, although lacking catalytic activity themselves, can interact with A′ and B′ to assemble into a catalytic complex that results in formation of E′. This does occur to some extent, as evidenced by the slight enhancement of exponential amplification in the presence compared to the absence of 0.5 μM each of A1 and A2-B (Supplementary Figure 3). Fortunately, this mechanism for spontaneous initiation of cross-replication is far less efficient compared to ligand-dependent initiation that proceeds through formation of E0. Spontaneous initiation can be made less efficient by reducing the stability of the A1•A2-B•A′ •B′ complex, but this also reduces the stability of the desired Etheo•A1•A2-B complex. The unligated components of E0 do not interact with A and B to form E, presumably because the stability of the A1•A2-B•A•B complex is much lower.
An analogous mechanism for spontaneous initiation of cross-replication can occur through assembly of an A•B•A′ •B′ complex that results in formation of E and/or E′. Efforts were made to prevent this from occurring, first by destabilizing the base-pairing interactions between A and A′, and second by reducing the concentrations of the four substrates. In two previous studies,8,9 the 3′ ends of A and A′ had the complementary sequences 5′-GAAU-3′ and 5′-GUUU-3′, respectively. Even this weak interaction enabled spontaneous initiation of amplification in a reaction mixture containing 5 μM each of A, A′, B, and B′. When the complementary sequences were shortened to 5′-GAU-3′ and 5′-GUU-3′, respectively (Figure 2b), the time to reach the threshold concentration of products increased from 2.0 to 4.2 h (Supplementary Table 1). When the concentrations of the four substrates were reduced from 5 to 1 μM each, the time to threshold increased to 8.6 hours, which is negligible compared to spontaneous initiation through the mechanism described above.
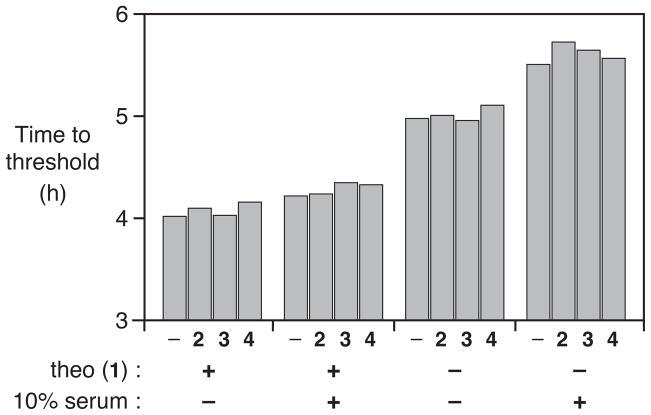
The theophylline aptamer is known to be highly specific for its cognate ligand.12 Not surprisingly, therefore, the addition of 200 μM concentration of various closely-related compounds, such as caffeine (2), theobromine (3), or 1,3-dimethyluric acid (4), had no effect on theophylline-dependent exponential amplification (Figure 5). This was found to be the case in either the absence or presence of 10% deproteinized serum. Similarly, the addition of 200 μM 2, 3, or 4 had no influence on the spontaneous initiation of amplification in the absence of the theophylline. Thus the dynamic range for the measurement of theophylline concentration is expected to be similar for a variety of complex samples.
Figure 5.
Detection of 200 μM theophylline (1), either alone or in the presence of 200 μM caffeine (2), theobromine (3), or 1,3-dimethyluric acid (4), measured in either the presence or absence of 10% human serum. The time to reach threshold was determined as shown in Figure 4.
Discussion
The previously described system of autocatalytic aptazymes enables the isothermal, ligand-dependent, exponential amplification of RNA.9 An important limitation of that system, however, is that the same RNA must both sense the ligand and serve as a substrate for the synthesis of new enzymes. The concentration regimes for these two functions may not be compatible when the concentration of the target ligand is below the Km of the enzyme. The present system avoids this limitation by separating the ligand-recognition and amplification functions, employing a ligand-dependent RNA ligase enzyme to generate a seed concentration of replicating ligase enzymes, which then initiate ligand-independent exponential amplification. This enables the quantitative detection of ligands in a concentration regime that is determined by the Kd of the aptazyme-ligand interaction. Employing an aptazyme that specifically recognizes theophylline, it was possible to measure theophylline concentrations over a saturation range of 0.06–0.84, based on the time required for exponential amplification to reach a defined threshold.
Other approaches have been taken to measure theophylline concentrations using the theophylline aptamer. In one study, a 2-aminopurine residue was introduced at a bulged position of the aptamer, where it is exposed to solvent and fluoresces in the absence of the ligand, but upon ligand binding forms base-stacking interactions that quench its fluorescence.14 The fluorescent signal can be used to determine the theophylline association rate or fraction bound at equilibrium, either of which can be used to measure theophylline concentration. In another study, a fluorescent nucleotide analogue was incorporated at a unique position within the aptamer, and the increase in fluorescence intensity that occurs upon theophylline binding was monitored.15 A third study employed an electrochemical detection method, tethering a ferrocene-labeled aptamer to a gold electrode and measuring the enhanced voltammetric response that occurs when theophylline is bound and the aptamer adopts a more compact folded state.16 All three of these approaches do not involve molecular amplification, but demonstrate the versatility of aptamer-based detection and its amenability to read-out by various methods.
The system described here for ligand recognition coupled to ligand-independent exponential amplification is modular in that amplification can be triggered by a variety of ligand-dependent seeding reactions that are linked to a common read-out method for monitoring the yield of amplified products over time. The present study employed radiolabeled materials that were analyzed by polyacrylamide gel electrophoresis, while a previous study relied on a luciferase reporter assay that was driven by inorganic pyrophosphate released during RNA ligation.9 For high-throughput applications it would be preferable to use fluorescently labeled substrates that are either captured or analyzed in situ using FRET, as commonly practiced with LCR.17,18 Like LCR, the amplification system described here can be multiplexed, with each ligand-dependent reaction seeding a different cross-replication reaction. Several distinct replicators can operate in parallel because the interaction between the replicating enzymes and their complementary substrates is highly sequence-specific.8
An important drawback of the ligand-dependent exponential amplification system is that it employs RNA molecules, which are susceptible to degradation by ribonucleases. Nucleases can be removed from the sample prior to analysis, but this will be problematic if the target ligand also is a protein. For such cases it will be necessary to develop nuclease-resistant forms of the cross-replicating RNA enzymes that contain 2′-O-methyl nucleotide analogs or other backbone modifications, as are commonly employed in therapeutic nucleic acids.19–25
Another limitation of the system is that it requires a few hours for exponential growth to reach the threshold concentration due to the slow catalytic rate of the RNA enzymes. In the present study the goal was to maximize the dynamic range of the assay, rather than minimize the time to threshold. Accordingly, the substrates were at sub-saturating concentrations and a form of the enzyme was used that is approximately 10-fold slower than the standard version,8 but is less susceptible to ligand-independent amplification. For applications in clinical diagnostics that do not require such a broad dynamic range, one could employ more reactive enzymes. The therapeutic range for theophylline, for example, is a serum concentration of 50–100 μM, which is at the upper end of the dynamic range explored in this study (Figure 4). Ultimately, one would want to employ RNA enzymes that are both highly reactive and not susceptible to ligand-independent amplification. This will require further enzyme engineering, likely involving a combination of rational design and directed evolution.
Conclusions
PCR and related amplification technologies have revolutionized molecular diagnostics by providing highly sensitive and specific methods for the detection of target nucleic acids. In qPCR, primer binding and extension on a target nucleic acid generates DNA molecules that seed target-independent exponential amplification. Analogously, RNA aptamers can be used to recognize various non-nucleic-acid targets, giving rise to RNA enzymes that seed exponential amplification. The amount of time required for amplification to reach a threshold concentration can be related to the concentration of the seed molecules, which is determined by the concentration of the target. Unlike qPCR, the RNA-based system operates at constant temperature and does not require any protein enzymes, but it also has the disadvantages that RNA molecules are susceptible to nuclease degradation and are slower catalysts compared to their protein counterparts. For some applications these disadvantages will be outweighed by the generality and high specificity of ligand-aptamer interactions.
Experimental section
Materials
Oligodeoxynucleotides templates for in vitro transcription were synthesized, purified, and desalted as described previously.9 Histidine-tagged T7 RNA polymerase and Thermus aquaticus DNA polymerase were expressed and purified as described previously.9,26 M1 RNA, the catalytic subunit of RNAse P, was prepared as described previously.8 Nucleoside and deoxynucleoside 5′-triphosphates, theophylline, theobromine, and 1,3-dimethyluric acid were purchased from Sigma-Aldrich (St. Louis, MO), [γ-32P]ATP (7 μCi/pmol) was from Perkin Elmer (Waltham, MA), and human serum (off-clot) and caffeine were from MP Biomedicals (Solon, OH).
Preparation of RNA enzymes and substrates
All RNA enzymes and substrates were prepared by in vitro transcription as described previously,8,9 except that for A2-B, B, and B′ the T7 φ2.5 promoter was used to increase 5′-terminal homogeneity and a self-cleaving hammerhead ribozyme was installed at the 3′ end to ensure 3′-terminal homogeneity. The hammerhead ribozymes had the sequences: 5′-CAACUUA•UCCGACGGAAACGUCGGACUGA-UGAGGCCGAAAGGCCGAAAAGUUG-3′, 5′-CAACUUA•UACGGAAACGUACUGAUGAGGCC-GAAAGGCCGAAAAGUUG-3′, and 5′-GAAUAUUC•UACGGAAACGUACUGAUGAGGCCGAA-AGGCCGAAAAUAUUC-3′ for A2-B, B, and B′, respectively (stem III pairing underlined, dot indicates the cleavage site). A1, A, and A′ were transcribed with appended 3′-terminal regions that were cleaved by E. coli M1 RNA to give precise 3′ ends.8 The external guide RNA27 for directing cleavage of A1 and A had the sequence 5′-CGUAAGUUGCGGUCUCACCA-3′, and for directing cleavage of A′ had the sequence 5′-AUAUUCAUGCGGUCUCACCA-3′ (hybridization region underlined). In order to reduce theophylline-independent amplification, the P1 stem of the RNA enzymes was shortened by one base pair compared to that employed in previous studies.8,9
Ligand-dependent catalysis
The reactions used to generate E0 employed 5 μM Etheo, 0.5 μM [5′-32P]-labeled A1, 0.5 μM A2-B, 25 mM MgCl2, 50 mM EPPS (pH 8.5), and various concentrations of theophylline, which were incubated at 42 °C for 20 or 30 min. Some reaction mixtures also contained 10% human serum that had been deproteinized by phenol extraction. Aliquots were removed from the reaction mixture and analyzed by denaturing polyacrylamide gel electrophoresis (PAGE), measuring the fraction of labeled A1 that had been converted to labeled product. The yield of E0 as a function of theophylline concentration follows a saturation plot: , where ε is the yield in the absence of theophylline and [E0]max is the yield at saturating theophylline concentration. The yield of E0 at time t was normalized to [E0]max to obtain the fractional saturation: .
Exponential amplification
The products of ligand-dependent catalysis were mixed with lyophilized substrates to initiate cross-catalytic exponential amplification, employing a final concentration of 1 μM each of A and B and 1.5 μM each of A′ and B′. The concentrations of A′ and B′ were 0.5 μM higher to compensate for complementary A1 and A2-B molecules that were carried over from the ligand-dependent reaction. Both A and A′ were [5′-32P]-labeled, and the reaction mixture was maintained at 42 °C. Aliquots were taken at various times and the amount of newly-synthesized E was determined by PAGE analysis. These data were fit to the logistic growth equation: , where a is the final extent, b is the degree of sigmoidicity, and c is the exponential growth rate.
Supplementary Material
Acknowledgments
This work was supported by Grant No. GM065130 from the National Institutes of Health.
Footnotes
Supporting Information Available. Three figures, showing the rate of Etheo as a function of theophylline concentration, quantitative theophylline-dependent amplification in the presence of 10% human serum, and the effect of adding either Etheo or a combination of A1 and A2-B to the amplification mixture; and one table comparing the spontaneous initiation of cross-replication under various reaction conditions. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Syvanen AC, Bengtstrom M, Tenhunen J, Soderlund H. Nucleic Acids Res. 1988;16:11327–11338. doi: 10.1093/nar/16.23.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang AM, Doyle MV, Mark DF. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sano T, Smith CL, Cantor CR. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 4.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, Östman A, Landegren U. Nat Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 5.Wu DY, Wallace RB. Genomics. 1989;4:560–569. doi: 10.1016/0888-7543(89)90280-2. [DOI] [PubMed] [Google Scholar]
- 6.Barany F. Proc Natl Acad Sci USA. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guatelli JC, Whitfield KM, Kwoh DY, Barringer KJ, Richman DD, Gingeras TR. Proc Natl Acad Sci USA. 1990;87:1874–1878. doi: 10.1073/pnas.87.5.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lincoln TA, Joyce GF. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam BJ, Joyce GF. Nat Biotechnol. 2009;27:288–292. doi: 10.1038/nbt.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riedy MC, Timm EA, Jr, Stewart CC. BioTechniques. 1995;18:70–74. [PubMed] [Google Scholar]
- 11.Klussmann S. The aptamer handbook: functional oligonucleotides and their applications. Wiley-VCH; Weinheim, Germany: 2006. [Google Scholar]
- 12.Jenison RD, Gill SC, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 13.Rogers J, Joyce GF. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jucker FM, Phillips RM, McCallum SA, Pardi A. Biochemistry. 2003;42:2560–2567. doi: 10.1021/bi027103+. [DOI] [PubMed] [Google Scholar]
- 15.Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S, Hirao I. J Am Chem Soc. 2005;127:17286–17295. doi: 10.1021/ja0542946. [DOI] [PubMed] [Google Scholar]
- 16.Ferapontova EE, Olsen EM, Gothelf KV. J Am Chem Soc. 2008;130:4256–4258. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Kwok PY. Genet Anal. 1999;14:157–163. doi: 10.1016/s1050-3862(98)00016-3. [DOI] [PubMed] [Google Scholar]
- 18.Wabuyele MB, Farquar H, Stryjewski W, Hammer RP, Soper SA, Cheng YW, Barany F. J Am Chem Soc. 2003;125:6937–6945. doi: 10.1021/ja034716g. [DOI] [PubMed] [Google Scholar]
- 19.Inoue H, Hayase Y, Imura A, Iwai S, Miura K, Ohtsuka E. Nucleic Acids Res. 1987;15:6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sproat BS, Lamond AI, Beijer B, Neuner P, Ryder U. Nucleic Acids Res. 1989;17:3373–3386. doi: 10.1093/nar/17.9.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paolella G, Sproat BS, Lamond AI. EMBO J. 1992;11:1913–1919. doi: 10.1002/j.1460-2075.1992.tb05244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green LS, Jellinek D, Bell C, Beebe LA, Feistner BD, Gill SC, Jucker FM, Janjic N. Chem Biol. 1995;2:683–695. doi: 10.1016/1074-5521(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 23.Monia BP, Johnston JF, Sasmor H, Cummins LL. J Biol Chem. 1996;271:14533–14540. doi: 10.1074/jbc.271.24.14533. [DOI] [PubMed] [Google Scholar]
- 24.Beaudry A, DeFoe J, Zinnen S, Burgin A, Beigelman L. Chem Biol. 2000;7:323–334. doi: 10.1016/s1074-5521(00)00110-1. [DOI] [PubMed] [Google Scholar]
- 25.Dupont DM, Madsen JB, Hartmann RK, Tavitian B, Ducongé F, Kjems J, Andreasen PA. RNA. 2010;16:2360–2369. doi: 10.1261/rna.2338210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pluthero FG. Nucleic Acids Res. 1993;21:4850–4851. doi: 10.1093/nar/21.20.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forster AC, Altman S. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.