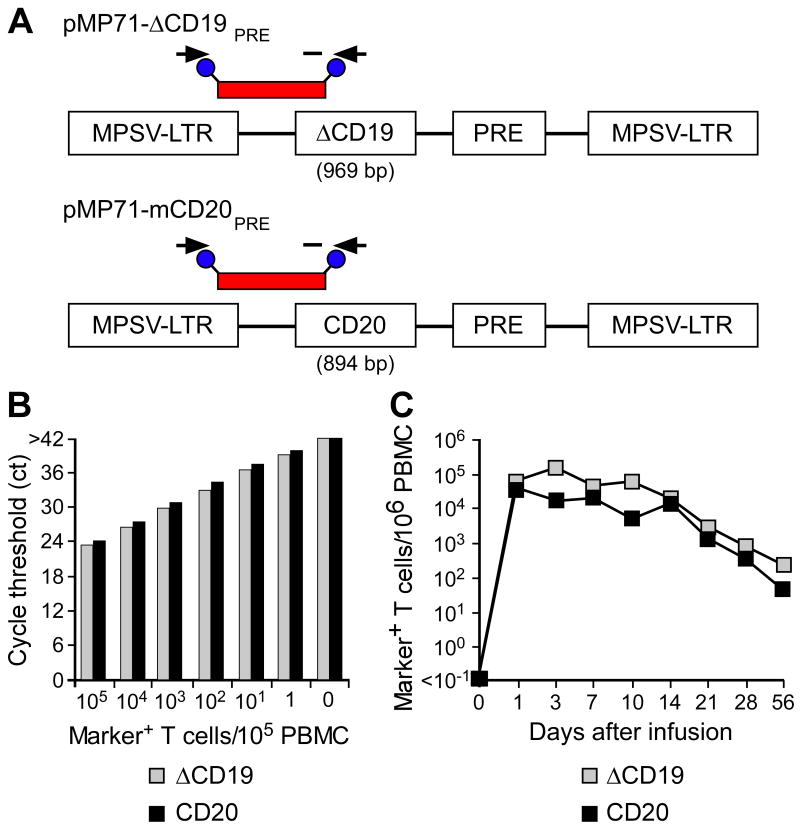
Fig. 4. Tracking of ΔCD19+ or CD20+ CMV-specific CD8+ TE clones following adoptive transfer by qPCR.
(A) Schematic design of retroviral vector constructs encoding for macaque B-cell lineage marker genes and location of primer and fluorescent probe (red bar) used for the qPCR assay. Abbreviations: MPSV-LTR, myeloproliferative sarcoma virus retroviral long terminal repeat; PRE, woodchuck hepatitis virus post-transcriptional regulatory element. (B) Detection of ΔCD19 or CD20-marked T cells within PBMC by qPCR. Samples of titrated numbers of ΔCD19+ ( ) or CD20+ T cells (■) were spiked into aliquots of pre-infusion PBMC (each 105 PBMC/reaction) and examined by real-time qPCR for detection of marker-positive T cells. Data are representative of each 3 assays with ΔCD19+ or CD20+ T cells. (C) Enumeration of transferred ΔCD19+ or CD20+ T cells determined by real-time qPCR for vector sequences. Autologous ΔCD19+ (
) or CD20+ T cells (■) were spiked into aliquots of pre-infusion PBMC (each 105 PBMC/reaction) and examined by real-time qPCR for detection of marker-positive T cells. Data are representative of each 3 assays with ΔCD19+ or CD20+ T cells. (C) Enumeration of transferred ΔCD19+ or CD20+ T cells determined by real-time qPCR for vector sequences. Autologous ΔCD19+ ( ) or CD20+ (■) TCM-derived TE clones were expanded in vitro and transferred to each one of the macaques at a dose of 5×108/kg. DNA was isolated from samples of PBMC obtained before and at indicated time-points after the T-cell infusion and examined by real-time qPCR for detection of marker-positive T cells.
) or CD20+ (■) TCM-derived TE clones were expanded in vitro and transferred to each one of the macaques at a dose of 5×108/kg. DNA was isolated from samples of PBMC obtained before and at indicated time-points after the T-cell infusion and examined by real-time qPCR for detection of marker-positive T cells.