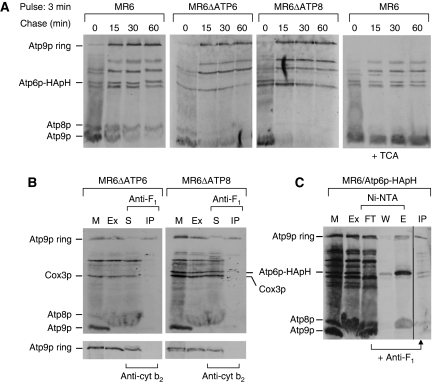
Figure 8.
Kinetics of Atp9p ring oligomerization and detection of an F1/Atp9p ring complex (A) Mitochondria were prepared from the respiratory competent parental strain MR6, from MR6ΔATP6, a mitochondrial mutant with an atp6 null allele and MR6ΔATP8/ATP6-HApH, a mitochondrial mutant with an atp8 null allele expressing Atp6p doubly tagged with HA and polyhistidine. Mitochondria (0.5 mg) were pulse labelled for 3 min (time 0), and were chased for 15, 30 and 60 min after addition of puromycin and excess cold methionine plus cysteine as in Figure 3. Total mitochondrial proteins were separated by SDS–PAGE on 17.5% polyacrylamide gel, transferred to nitrocellulose and exposed to the X-ray film. In the extreme right hand panel, the same samples of MR6 shown in the left panel were treated with 5% TCA before depolymerization in sample buffer. The conversion of the Atp9 ring to the monomer by this treatment increases the transfer efficiency causing a significant enhancement of the signal. (B) Mitochondria (1 mg) of MR6ΔATP6 and MR6ΔATP8/ATP6-HApH were labelled for 20 min, extracted with digitonin and the extracts were incubated with polyclonal antibody against F1 or cytochrome b2 for 1 h at room temperature. The immunoprecipitates were removed by centrifugation at 14 000 r.p.m. for 15 min. Mitochondria (M), the digitonin extract (Ex), the supernatant from the antibody precipitate (S) and the antibody precipitate (IP) were analysed by SDS–PAGE as in (A). (C) Mitochondria of MR6/ATP6-HApH were labelled for 20 min, extracted with 3% digitonin and purified on Ni-NTA. The protein fraction that was not adsorbed to the Ni-NTA beads was incubated with antibody against F1 and the resultant antibody precipitate (IP) was analysed by SDS–PAGE as in (B). All the lanes shown are part of the same gel exposed for the same period of time.