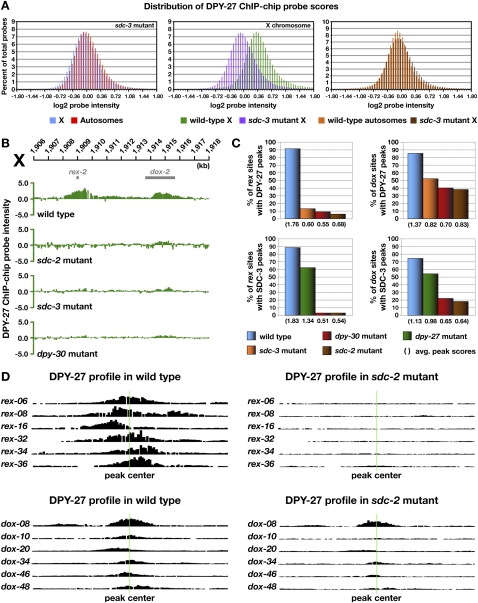
Figure 3.
Differential requirements for DCC binding to rex and dox sites. (A) Histograms show distribution of DPY-27 ChIP–chip probe scores across X and autosomes in wild-type versus sdc-3 mutant embryos. Probe scores were analyzed as in Figure 2D. The probe score distribution for X in sdc-3 mutant embryos is greatly reduced compared with that in wild-type embryos and closely resembles that of wild-type autosomes (see Fig. 2D). Additional histograms of probe intensities from DPY-27 ChIP–chip experiments for other DCC mutants are in Supplemental Figure S4. (B) ChIP–chip profile of DPY-27 binding in wild-type XX embryos compared with embryos mutant for dpy-30, sdc-3, or sdc-2. rex site binding is nearly eliminated in these mutants, while dox site binding is greatly reduced but present. (C) Histograms showing quantification of DPY-27 and SDC-3 binding at rex and dox sites in wild-type versus DC mutant embryos. The height of the bar corresponds to the percentage of bound rex and dox sites with a peak score of 0.75 or better in DC mutants. The average peak scores shown in parentheses below the histogram were calculated from the scores of all called peaks at rex and dox sites. DCC binding to rex sites is dependent on SDC-2, SDC-3, and DPY-30, whereas a low-level dox site binding is independent of these proteins. (D) Graphical representations of DPY-27 ChIP–chip probe intensities along 5-kb regions centered on representative rex and dox sites (green line) in wild-type versus sdc-2 mutant embryos further show that binding to rex sites is eliminated and binding to dox sites is severely reduced. Additional graphical representations of probe intensities from SDC-3 and DPY-27 ChIP–chip experiments for other DCC mutants are in Supplemental Figure S5.