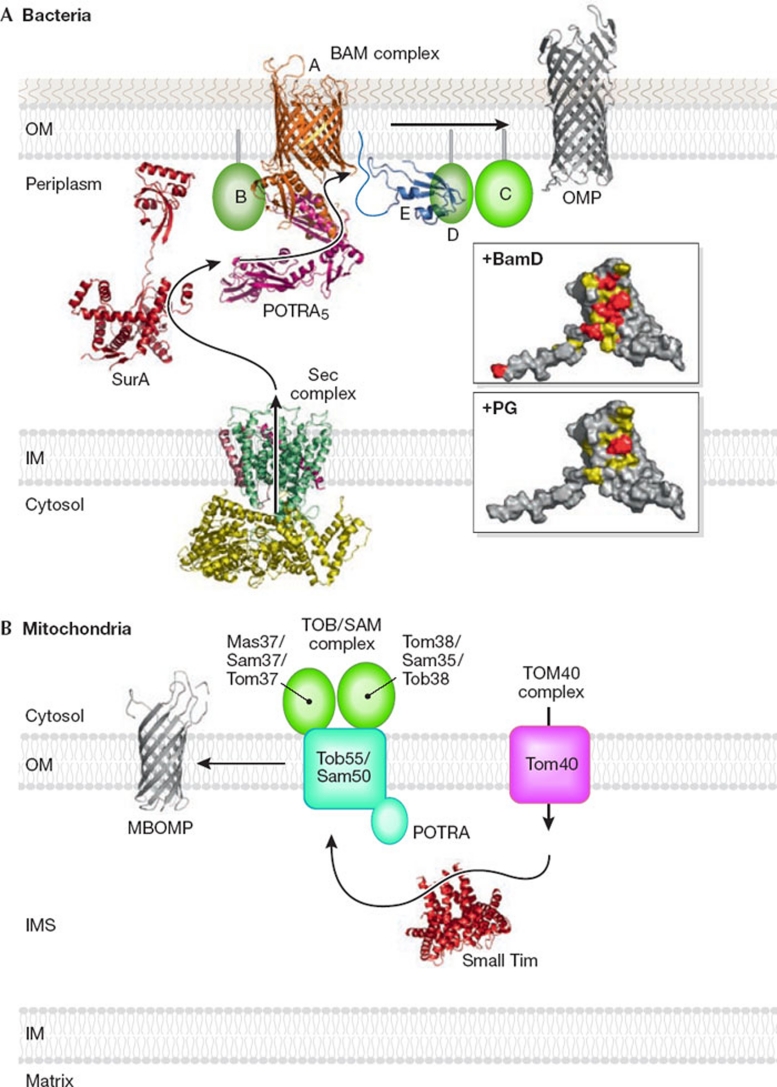
Figure 1.
β-barrel protein assembly in bacterial and mitochondrial outer membranes. (A) Bacteria. Ribbon models of the structures of the Sec complex, SurA, BamA (Clantin et al, 2007; Kim et al, 2007), BamE and OMP. The upper and lower inserts show the surface of BamE (residues 20–108; viewed after approximately 90° rotation of the ribbon model around the horizontal axis toward the reader). Residues important for BamD binding are shown in red and residues with NMR signals that were perturbed by BamD binding are shown in yellow. The residue (Phe 74) important for PG binding is shown in red and the residues with NMR signals that were perturbed by PG binding are shown in yellow. (B) Mitochondria. Ribbon models were drawn for the structures of small Tim and MBOMP. IM, inner membrane; IMS, intermembrane space; MBOMP, mitochondrial β-barrel outer membrane protein; OM, outer membrane; OMP, outer membrane protein; PG, phosphatidylglycerol; POTRA, polypeptide transport-associated domain.