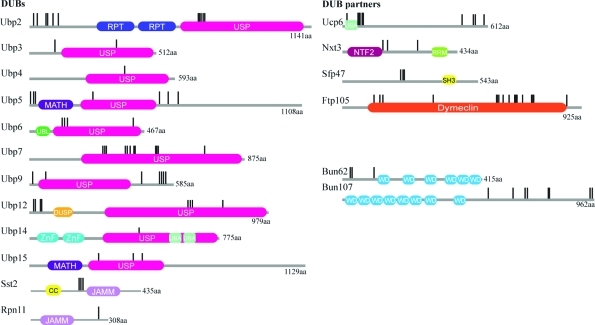
Figure 3.
Domain architecture and mapping of detected phosphorylation sites within the S. pombe DUBs and their partners. Domain architectures were retrieved using the SMART and Pfam databases. The following domains were found: USP (ubiquitin-specific proteases) JAMM (JAB1/MPN/Mov34 metalloenzymes), DUSP (Domain in Ubiquitin-specific proteases), MATH (Meprin and TRAF homology), UBL (Ubiquitin-like), ZnF (Ubiquitin Carboxyl-terminal hydrolase-like zinc finger), UBA (Ubiquitin-associated). RPT are internal repeats. Phosphosites are denoted by vertical black lines.