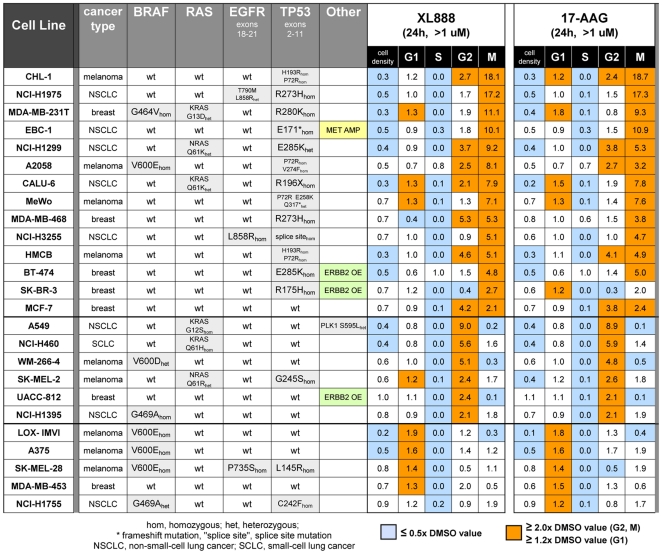
Figure 3. Cell cycle analysis of 25 cancer cell lines treated with XL888 or 17-AAG.
Cells were treated with 1–1.6 uM XL888 or 17-AAG for 24 h, and cell cycle profiles were analyzed by the HC cell cycle method. Cell cycle data is normalized to the DMSO value for a given phase and given cell line and is represented as a fold-change vs. DMSO. The heat map color key is as follows: light blue, ≤0.5× DMSO value; orange, ≥2× DMSO value for G2 and M and ≥1.2× DMSO value for G1. Data is successively sorted in descending order of (1) accumulation in M, (2) accumulation in G2, and (3) accumulation in G1. Mutations are highlighted in gray. EBC-1 is a MET-amplified line (“MET AMP”) and BT-474, UACC-812, and SK-BR-3 are ERBB2-overexpessing lines (“ERBB2 OE”). Genotype data in this figure is derived from COSMIC [75] or from in-house sequencing. See Figure S1B for a version of the heat map that includes apoptosis and proliferation data as well as cell cycle profile data. Figure S1B also shows cell cycle phenotypes of cells treated with 0.4 uM XL888 or 17-AAG for 24 h, in comparison to the 1.1–1.67 treatment shown here. Chi-square analysis indicates that the probability of the observed correlation of mutant p53 lines with M+/−G2 status being random is 0.0089; see legend to Figure S1 for details.