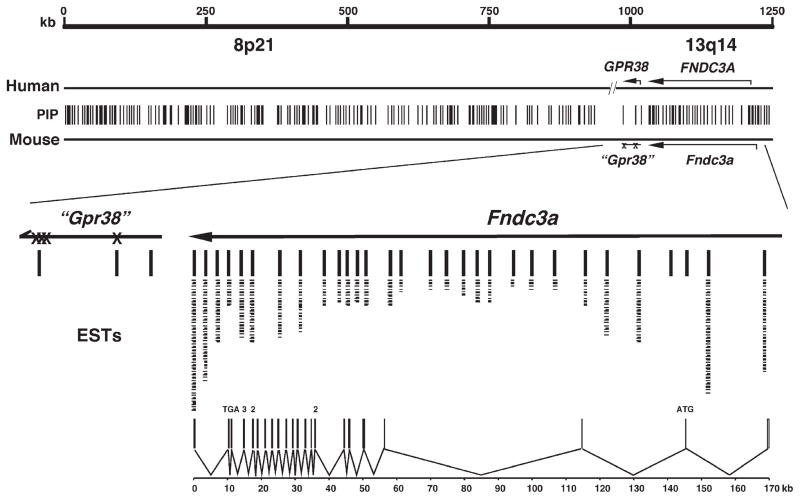
Fig. 3.
Comparative genome analysis of the 1.2 Mb region of chromosome 14 deleted in sys mice with human 8p21 and 13q14 suggests that Fndc3a is the only gene mutated in sys mice. The scale shown is for mouse chromosome 14. Vertical lines (PIP) represent the 223 pairwise alignments of conserved DNA (>75% identity over >100 bp) as identified using PipMaker (http://pipmaker.bx.psu.edu/pipmaker/). Of the 223 conserved regions, only those within the Fndc3a locus had identity to mouse ESTs as determined by BLAST analysis. Individual ESTs are represented by a dash under the conserved regions. Lines are used to indicate identity between Fndc3a exons and the respective ESTs and conserved regions. Two regions at the 5′ end of Fndc3a are conserved between human and mouse but did not identify an EST in the database. These sequences are not found in the predicted full-length Fndc3a cDNA, have no ORF, and may therefore represent control elements for this locus. (X)—denotes nonsense mutations within the region of mouse chromosome 14 (“Gpr38”) with overall similarity to human GPR38. The lower part of the figure depicts the structure of the Fndc3a mouse gene, composed of 27 exons distributed over 170 kb (PIP—percent identity plot; Mb—megabase pairs; EST—expressed sequence tag; ORF—open reading frame).