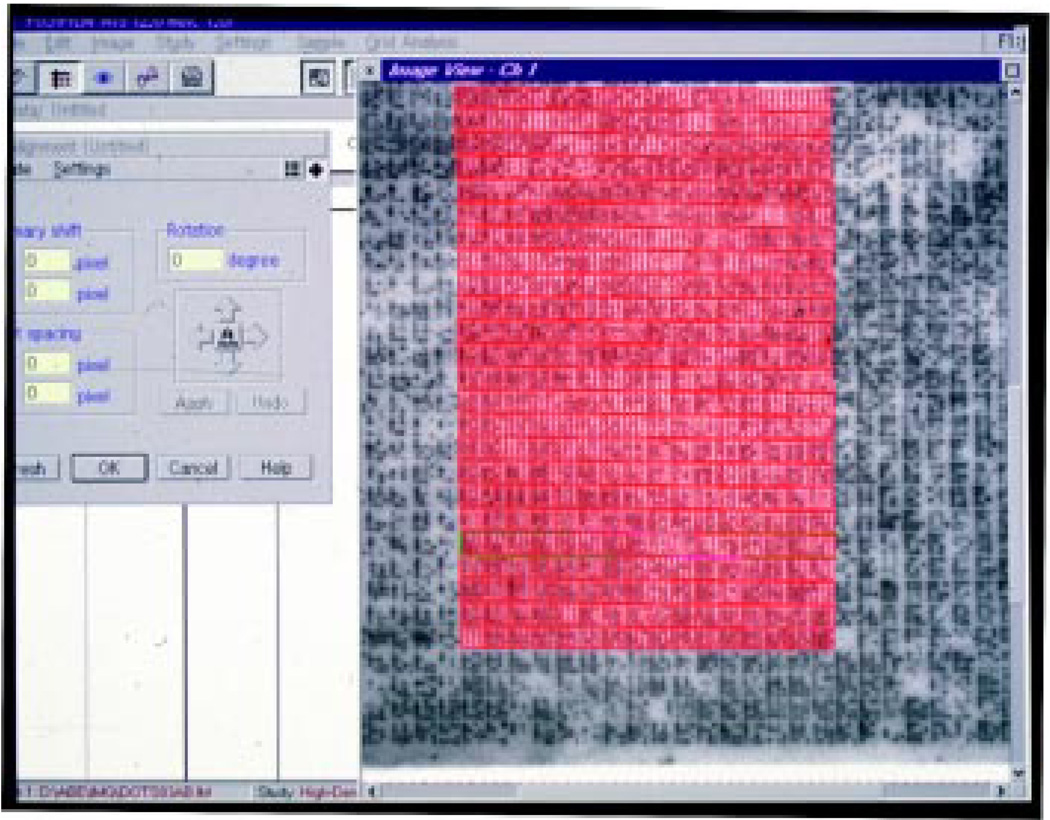
Fig. 4. Use of AIS software to quantitate relative levels of gene expression.
A 22×22cm hybridization filter containing approximately 36864 (4×4×384) individual mouse genomic DNA BAC clones spotted in 4×4 arrays was hybridized with a complex probe, washed and exposed to a phospho-storage screen. The plate was read by a phospho-imager (Fuji BAS 2000) and the image directly imported by the AIS software program. The screen-shot illustrates a grid used by the software to define individual areas that are subjected to relative quantitation. The grid represents one sixth of the entire filter. The software locates spots, quantitates the signals and converts them into numerical data that can be imported to any spread-sheet application. Signals are normalized to a standard reference set of target sequences on the filter. The AIS software can also compare one hybridization ‘signature’ with another and present the results in either a numerical or graphic format. More recently, EXPANEL software has been developed to perform relative quantitation of hybridization signatures (M.S.H.K., unpublished). The EXPANEL application provides a 2-dimensional color-graded graphical representation of differential expression of addressed cDNAs.