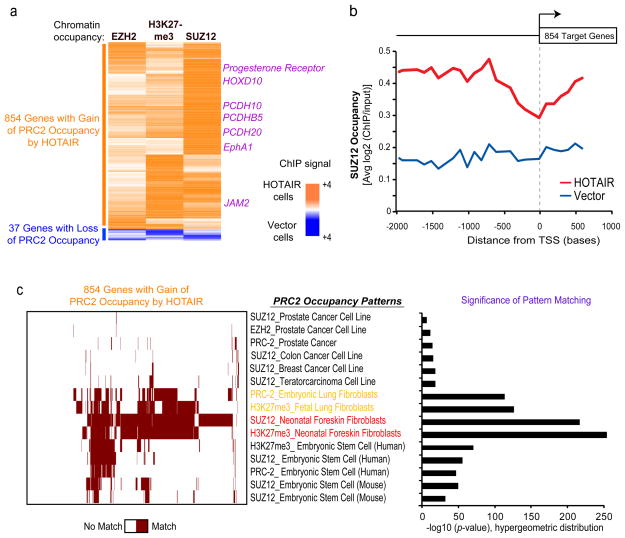
Figure 3. HOTAIR promotes selective re-targeting of PRC2 and H3K27me3 genome-wide.
a, Heat map representing genes with a significant relative change in chromatin occupancy of EZH2, SUZ12, and H3K27 following HOTAIR expression. MDA-MB-231 vector or HOTAIR cells were subjected to ChIP using anti-EZH2, H3K27me3, and SUZ12 antibodies followed by interrogation on a genome-wide promoter array. Values are depicted as relative ratio of HOTAIR over vector cells and represented as an orange-blue scale. b, Average SUZ12 occupancy of >800 PRC-2 target genes in HOTAIR or vector expressing cells across the length of gene promoter and gene body. All target genes are aligned by their transcriptional start sites (TSS). c, Module map20 of the 854 genes with a gain in PRC2 occupancy following HOTAR overexpression. (Left panel) Heat map of genes (column) showing a gain in PRC2 occupancy following HOTAR expression in breast carcinoma cells [see panel (a)] compared with PRC-2 occupancy patterns from the indicated cell or tissue type (rows). Binary scale is brown (match) or white (no match). (Right panel) Quantification of significance of pattern matching between gene sets.