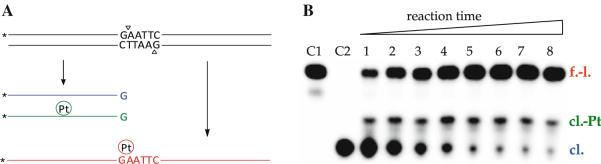
Fig. 2.
a The cleavage inhibition assay. Incubations of the platinum-treated 40-base-pair duplex (black) with EcoRI and subsequent separation of the mixtures by denaturing polyacrylamide gel electrophoresis (PAGE) results in three detectable products: unmodified (blue) and platinum-modified (green) 18-base-pair fragments, and uncleaved top strand (red). The asterisks denote the 32P label, and the triangles indicate the sites of endonucleolytic strand scission. b Representative denaturing polyacrylamide gel (12% acrylamide, 8 M urea) for the cleavage inhibition assay performed with compound 2. Lane assignments: C1 control—unmodified probe sequence; C2 control—unmodified, EcoRI-digested sequence; 1–8 reactions with complex 2 quenched after 20, 40 min, 1, 1.5, 2.5, 3.5, 4.5, and 6 h, respectively, followed by EcoRI digestion. The bands are labeled cl. for the 18-base-pair fragment, cl.-Pt for the platinum-containing 18-base-pair fragment, and f.-l. for the full-length fragment