Fig 5.
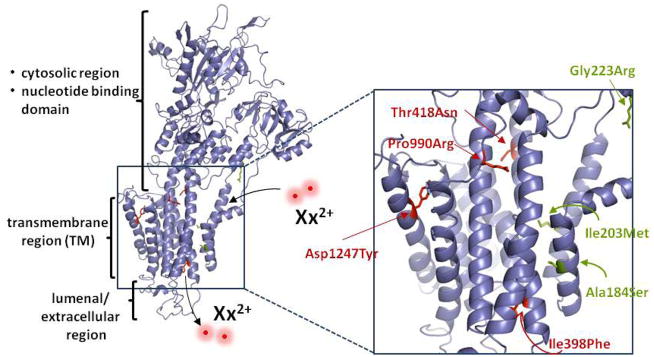
Resistance-associated SNPs map to the predicted transmembrane region of PfATP4. A homology model of PfATP4 was generated in SWISS-MODEL based on the crystal structure of the rabbit SERCA pump. Amino acid alignment analysis by EMBOSS (41) revealed 30% identity and 48% similarity between these proteins. Residues corresponding to resistance-associated mutations are indicated in red for NITD609-RDd2 and in green for NITD678-RDd2. These mutations mapped to the putative transmembrane helices. The sites of divalent cation entry and exit are indicated as Xx2+.