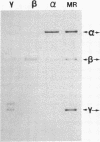
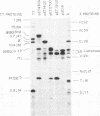
Abstract
The genes that encode the α, β, and γ subunits of component C of methyl coenzyme M reductase (mcrA, mcrB, and mcrG) in Methanococcus vannielii have been cloned and sequenced, and their expression in Escherichia coli has been demonstrated. These genes are organized into a five-gene cluster, mcrBDCGA, which contains two genes, designated mcrC and mcrD, with unknown functions. The mcr genes are separated by very short intergenic regions that contain multiple translation stop codons and strong ribosomebinding sequences. Although the genome of M. vannielii is 69 mol% A+T, there is a very strong preference in the mcrA, mcrB, and mcrG genes for the codon with a C in the wobble position in the codon pairs AAUC (asparagine), GAUC (aspartic acid), CAUC (histidine), AUUC (isoleucine), UUUC (phenylalanine), and UAUC (tyrosine). The mcrC and mcrD genes do not show this codon preference and frequently have U or A in the wobble position. As the codon pairs listed above are likely to be translated by the same tRNA with a G in the first anticodon position, the presence of C in the wobble position might ensure maximum efficiency of translation of transcripts of these very highly expressed genes.
Keywords: methyl reductase, archaebacteria, DNA sequence, codon usage, translation regulation
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckler G. S., Reeve J. N. Conservation of primary structure in the hisI gene of the archaebacterium, Methanococcus vannielii, the eubacterium Escherichia coli, and the eucaryote Saccharomyces cerevisiae. Mol Gen Genet. 1986 Jul;204(1):133–140. doi: 10.1007/BF00330200. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Cue D., Beckler G. S., Reeve J. N., Konisky J. Structure and sequence divergence of two archaebacterial genes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4207–4211. doi: 10.1073/pnas.82.12.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels L., Sparling R., Sprott G. D. The bioenergetics of methanogenesis. Biochim Biophys Acta. 1984 Sep 6;768(2):113–163. doi: 10.1016/0304-4173(84)90002-8. [DOI] [PubMed] [Google Scholar]
- Ellefson W. L., Wolfe R. S. Component C of the methylreductase system of Methanobacterium. J Biol Chem. 1981 May 10;256(9):4259–4262. [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Gunsalus R. P., Wolfe R. S. Methyl coenzyme M reductase from Methanobacterium thermoautotrophicum. Resolution and properties of the components. J Biol Chem. 1980 Mar 10;255(5):1891–1895. [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Reeve J. Use of minicells for bacteriophage-directed polypeptide synthesis. Methods Enzymol. 1979;68:493–503. doi: 10.1016/0076-6879(79)68038-2. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Wackett L. P., Hartwieg E. A., King J. A., Orme-Johnson W. H., Walsh C. T. Electron microscopy of nickel-containing methanogenic enzymes: methyl reductase and F420-reducing hydrogenase. J Bacteriol. 1987 Feb;169(2):718–727. doi: 10.1128/jb.169.2.718-727.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wich G., Hummel H., Jarsch M., Bär U., Böck A. Transcription signals for stable RNA genes in Methanococcus. Nucleic Acids Res. 1986 Mar 25;14(6):2459–2479. doi: 10.1093/nar/14.6.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wich G., Jarsch M., Böck A. Apparent operon for a 5S ribosomal RNA gene and for tRNA genes in the archaebacterium Methanococcus vannielii. Mol Gen Genet. 1984;196(1):146–151. doi: 10.1007/BF00334107. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]