Figure 1. Platform to assess EGFR Phosphorylation using LC-MS/MS.
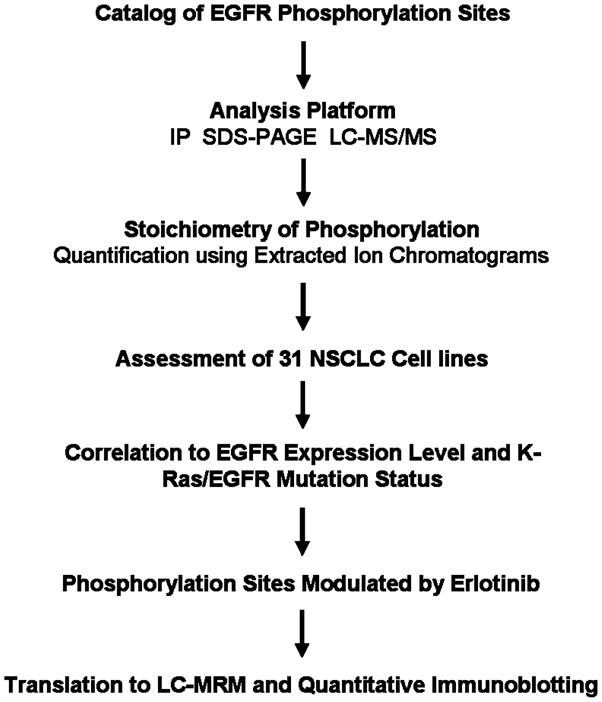
EGFR phosphorylation sites were determined using LC-MS/MS analysis of recombinant protein and EGFR extracted from HCC827 cells using SDS-PAGE fractionation and immunoprecipitation followed by SDS-PAGE. The latter sample preparation (IP) is used for all subsequent experiments. Stoichiometry is estimated using extracted ion chromatograms from high resolution LC-MS/MS. Phosphorylation is measured across a representative panel of cell lines, where each site can be correlated with mutations in EGFR or KRas and in lung cancer cells exposed to erlotinib, a EGFR tyrosine kinase inhibitor. Selected sites can be used for patient monitoring; examples are shown for translation to reaction monitoring mass spectrometry and quantitative immunoblotting, which are more amenable to analysis of patient samples.
