Abstract
The nuclear matrix bound transcription factor RUNX2 is a lineage-specific developmental regulator that is linked to cancer. We have previously shown that RUNX2 controls transcription of both RNA polymerase II genes and RNA polymerase I dependent ribosomal RNA genes. RUNX2 is epigenetically retained through mitosis on both classes of target genes in condensed chromosomes. We have used fluorescence recovery after photobleaching (FRAP) to measure the relative binding kinetics of EGFP-RUNX2 at transcription sites in the nucleus and nucleoli during interphase, as well as on mitotic chromosomes. RUNX2 becomes more strongly bound as cells go from interphase through prophase, with a doubling of the most tightly bound “immobile fraction”. RUNX2 exchange then becomes much more facile during metaphase to telophase. During interphase the less tightly bound pool of RUNX2 exchanges more slowly at nucleoli than at subnuclear foci, and the non-exchanging immobile fraction is greater in nucleoli. These results are consistent with a model in which the molecular mechanism of RUNX2 binding is different at protein-coding and ribosomal RNA genes. The binding interactions of RUNX2 change as cells go through mitosis, with binding affinity increasing as chromosomes condense and then decreasing through subsequent mitotic phases. The increased residence of RUNX2 at mitotic chromosomes may reflect its epigenetic function in “bookmarking” of target genes in cancer cells.
Keywords: Fluorescence Recovery After Photobleaching (FRAP), mitosis, epigenetics, RUNX2, transcription, gene expression, nuclear architecture, nuclear matrix, chromatin, chromosome, cell cycle, cancer
INTRODUCTION
The architectural organization of regulatory factors in the interphase nucleus and on mitotic chromosomes is functionally linked to biological control, as well as the onset and progression of tumorigenesis. RUNX/AML transcription factors represent a key class of lineage-specific gene regulators that associate with different micro-environments during the cell cycle. These proteins also trans-activate tissue-specific genes in the hematopoietic, neural, gastrointestinal, and bone developmental lineages. RUNX2 is a master regulator of skeletal development, exerting positive and negative control in the differentiation of both osteoblasts and chondrocytes, as well as tissue-specific gene expression (Lian et al, 2006;Komori, 2010).
RUNX2 was initially identified as a bone-specific nuclear matrix protein, NMP2, that interacts with the osteocalcin promoter (Bidwell et al, 1993;Merriman et al, 1995). Subsequent studies established that the localization of RUNX proteins to subnuclear foci requires a conserved nuclear matrix targeting signal (NMTS), which is present in RUNX1, RUNX2 and RUNX3 (Zeng et al, 1997;Zaidi et al, 2001;Pande et al, 2009). The NMTS supports binding of RUNX proteins to the nuclear matrix to spatially position these transcription factors in small structures that are distributed in a focal pattern throughout the nucleus (Zeng et al, 1997;Zeng et al, 1998). Some of these foci are sites of RNA polymerase II transcription. RUNX2, like other transcription factors, has a dynamic binding at these positionally stable sites with a half-time of exchange of about 10 seconds (Harrington et al, 2002). Loss of the NMTS has been linked to abrogated biological control during development (Choi et al, 2001;Dowdy et al, 2010) and in cancer (Bakshi et al, 2008).
More recently, we have identified RUNX2 at nucleoli and in nucleolar organizing regions where it represses RNA polymerase I dependent rDNA transcription (Young et al, 2007b). RUNX2 forms complexes with RNA polymerase I and the rDNA transcription factor Upstream Binding Factor (UBF1) (Jantzen et al, 1990;Pikaard et al, 1990;Ali et al, 2010). It can be found at rDNA promoters by ChIP and its binding at these promoters can result in epigenetic chromatin modifications and a decrease in rRNA synthesis (Drygin et al, 2010;Grummt and Voit, 2010;McStay and Grummt, 2008;Ali et al, 2010). Based on these findings, we have proposed that RUNX2 may coordinately regulate ribosome biogenesis and protein biosynthesis by modulating the activities of both RNA polymerases I and II (Young et al, 2007b).
RUNX2 represents the prototype for a new class of mitotic regulators with the dual function of supporting the biochemical requirements for transcription during interphase and preserving epigenetic control during mitosis by remaining associated with its target genes (Young et al, 2007b;Young et al, 2007a;Zaidi et al, 2010). Subsequently, we have shown that this novel epigenetic function first discovered for RUNX2 in osteoblasts is also mediated by RUNX3 in gastro-intestinal cells and by RUNX1 in the myeloid lineage, as well as by the t(8;21)-related fusion protein RUNX1/AML1-ETO in leukemia, reflecting compromised epigenetic control in cancer cells (Bakshi et al, 2008). Transcription factor mediated epigenetic control also operates in a broader biological context as evidenced by mitotic retention of myogenic and adipogenic transcription factors (e.g., myogenin and C/EBPbeta) during mesenchymal lineage progression (Ali et al, 2008). Thus, our studies suggest a common mechanism for coordinating rRNA and mRNA expression in different cell lineages.
At mitosis, RUNX2 remains associated with target genes on condensed chromosomes (Young et al, 2007a). While the NMTS is required for nuclear matrix association of RUNX proteins during interphase, DNA binding is essential for their interaction with mitotic chromosomes, which provides continuity of gene regulation and architectural organization in progeny cells. Mitotic targets include both RNA polymerase II protein coding genes and RNA polymerase I rRNA genes. We propose that RUNX2 functions to “bookmark” lineage-specific genes from the mother cell for expression in the daughter cells. This model is consistent with our previous studies showing that Myo D, Myogenin, CEBPbeta (Ali et al, 2008) and the leukemogenic t(8;21) fusion protein AML1-ETO (Bakshi et al, 2008) remain associated with target genes at mitosis. This “bookmarking” of genes by direct transcription factor association may maintain appropriate levels of phenotypic gene expression from one cell generation to the next (Zaidi et al, 2010).
The physical chemistry of macromolecular binding is readily studied in live cells by Fluorescence Recovery after Photobleaching (FRAP) and related photobleaching techniques that measure the binding or diffusion of fluorescent molecules introduced into a cell (Nickerson, 2009). After fluorescent molecules are photobleached, the “dark” molecules in the bleach zone are replaced by homologous fluorescent molecules from other regions of the cell. When the fluorescent molecule is bound in a relatively immobile complex (i.e., a complex that is immobile over the time course of the experiment measured in seconds to minutes), the mechanism for this replacement must include unbinding of the “dark” molecules in the bleach zone, the unbinding of still fluorescent molecules outside of the bleach zone, the exchange of these two pools of molecules by diffusion, and the binding of still fluorescent molecules to complexes in the bleach zone (Lele et al, 2004). Thus the rate of recovery depends on binding and unbinding constants, and on diffusion coefficients (Lele et al, 2004;Lele et al, 2006;Lele and Ingber, 2006;Carrero et al, 2003;Phair et al, 2004;Phair and Misteli, 2001;Sprague et al, 2006;Beaudouin et al, 2006). In most experiments with nuclear proteins, including all experiments we report here, rates of diffusion are much faster than binding and unbinding rates. These experiments, therefore, provide a measure of molecular exchange in complexes (Kruhlak et al, 2000;Nickerson, 2009).
To understand the cellular mechanisms that support the functions of epigenetic transcription factors like RUNX2 at mitosis, it is necessary to define their dynamic behavior in live cells by time-lapse fluorescence microscopy and FRAP analysis. In this study, we used FRAP to measure the binding affinity of RUNX2 to sites on mitotic chromosomes, at subnuclear sites, and at nucleoli. We find that RUNX2 is associated with chromosomes throughout mitosis, but that the kinetics of exchange at these sites change with mitotic progression. These data indicate molecular differences in the interactions of RUNX2 containing complexes with binding sites at different cell cycle stages.
MATERIALS AND METHODS
Cell culture and transfections
Human osteosarcoma cells (U2OS) were grown in McCoy's 5A media (Gibco, Carlsbad, CA) with 10% Fetal Bovine Serum (Atlanta Biologicals, Lawrenceville, GA), 1% L-Glutamine (Gibco) and 1% Penicillin-Streptomycin (Gibco). For live cell experiments, 3 × 105 cells were seeded on sterile 40 mm coverslips. The cells were then co-transfected with 300ng Enhanced Green Fluorescent Protein (EGFP)-RUNX2 and 200 ng mRFP- histone H2B plasmids using the transfection reagent FuGENE 6 (Roche, Indianapolis, IN) for 24 hours before observation or photobleaching. Standard procedures for cloning fluorescent fusion proteins were as previously described (Harrington et al, 2002;Wagner et al, 2003;Wagner et al, 2004;Kota et al, 2008).
Live cell microscopy
Cells seeded on 40 mm coverslips were observed at 37°C in a Bioptechs FCS2 Closed Chamber System using a Bioptechs objective heater. The chamber was assembled in a 37°C environmental chamber and media, supplemented with 20mM HEPES pH 7.5., was pre-warmed to 37°C and perfused across the coverslip.
Fluorescence recovery after photobleaching
FRAP assays were performed at 37°C as previously described (Wagner et al, 2003;Kota et al, 2008). Leica Confocal Software (Leica Microsystems, Exton, PA) was used to measure the intensity of fluorescence in the bleached region of interest and in the whole nuclear profile at each time point. We analyzed the data using Microsoft Excel spreadsheets. Any remaining fluorescence in the bleached area after bleach was normalized to zero. To calculate the relative fluorescence intensity (Irel) in the bleached area at time t, we used the equation: Irel, t = (It*(N0/Nt))−(Ipbl*(N0/Npbl))/(I0−(Ipbl*(N0/Npbl))). N0 is the total nuclear fluorescence before bleaching, Npbl is the total nuclear fluorescence in the first image taken after the bleach, Nt is the total nuclear fluorescence at time t, I0 is the fluorescence in the bleach zone before the bleach, It is the fluorescence in the bleach zone at time t, and Ipbl is the fluorescence in the bleach zone in the first image taken after the bleach. Curve-fitting was performed using Prism 5 (Graphpad Software). The best fit for these photobleach recoveries was obtained using an exponential association curve: F(t) = Fmax (1− e−kt). All half times of recovery and immobile fractions were calculated from a best fit to this equation. Individual time points are presented as means with error bars showing standard errors.
We observed a greater sensitivity of mitotic cells to 488 nm laser light after EGFP-RUNX2 transfection, sometimes resulting in mitotic arrest. To reduce the exposure of cells to light, mitotic cells were located and microscope parameters set by observing mRFP-H2B with 568 nm excitation light. Live cells were then imaged and subjected to photobleaching at 488 nm for FRAP analysis.
RESULTS
Retention of RUNX2 on mitotic chromosomes in live cells
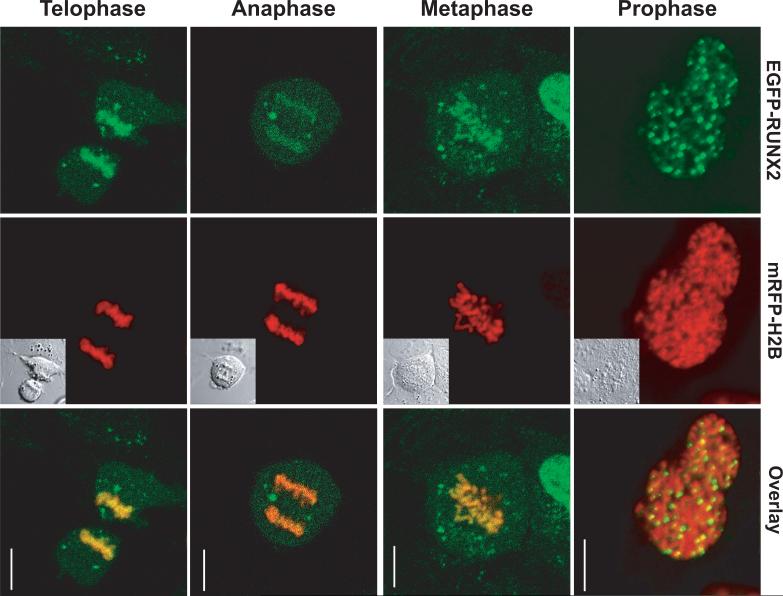
The epigenetic function of RUNX2 is linked to its retention at target genes on condensed chromosomes throughout mitosis (Zaidi et al, 2010). RUNX2 controls gene transcription through an interdependent cohort of co-regulatory factors that differentially associate with mitotic chromosomes (Zaidi et al, 2010;Ali et al, 2010). We used microscopic imaging to investigate whether RUNX2 remains bound to mitotic chromosomes in live cells and to quantify the dynamics of that epigenetically important association. U2OS cells transiently expressing EGFP-RUNX2 and mRFP-Histone H2B were examined in a live-cell chamber by confocal microscopy at 24 to 36 hours after transfection. The EGFP-RUNX2 fusion protein is fully functional in transcriptional activation (Harrington et al, 2002) and the labeled histone H2B facilitates the identification of mitotic stages. RUNX2 is localized at nuclear foci in interphase cells (Harrington et al, 2002) and retained on condensed chromosomes in cells at all stages of mitosis (Fig. 1). The movement of RUNX2 on chromosomes through transitions in mitosis was established by time-lapse imaging (Supplementary Data). The localization of RUNX2 in live cells as observed by conventional or confocal fluorescence microscopy (Figs. 1 and 2) is indistinguishable from its localization in fixed cells (Young et al, 2007a;Harrington et al, 2002).
Fig. 1. RUNX2 remains bound on mitotic chromosomes throughout mitosis.
U2OS human osteosarcoma cells were transfected with EGFP-RUNX2 and mRFP-Histone H2B. Mitotic cells were located using mRFP-Histone H2B and imaged through mitosis. RUNX2 punctate foci as well as diffuse RUNX2 staining on chromosomes were clearly evident in prophase, metaphase, anaphase and telophase. Left column—EGFP-RUNX2; middle column—mRFP-H2B and DIC in insert for comparison; right column—the merged image. Size bar is 10 μm.
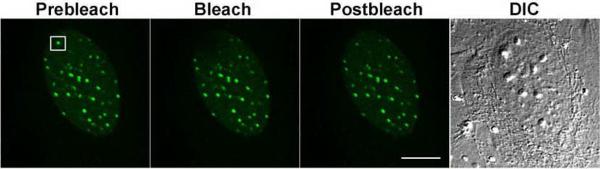
Fig. 2. RUNX2 binding kinetics in interphase non-nucleolar foci.
In interphase cells, RUNX2 subnuclear foci, sites of RNA polymerase II transcription, were photobleached for 2 seconds and recovery was monitored for 80–120 seconds. The Differential Interference Contrast (DIC) image is shown for comparison. The calculated half-time (t1/2) of recovery was 12 seconds and the immobile fraction was 35%. Size bar is 10 μm.
The kinetics of RUNX2 binding at target genes are different in interphase and during mitosis
RUNX2 is a transcriptional regulator of both RNA polymerase I genes at nucleoli and of RNA polymerase II genes in the nucleus (Young et al, 2007a), with spatially segregated gene regulatory activities and co-regulatory factor interactions. FRAP was used to assess whether there are differences in the equilibrium binding of RUNX2 that may control RNA polymerase I or RNA polymerase II mediated transcription. We compared the relative binding affinity and exchange of RUNX2 at chromosomal binding sites during mitosis or during interphase at RNA polymerase I related nucleolar sites and at RNA polymerase ll related non-nucleolar sites.
To understand the affinity of RUNX2 binding to chromatin within distinct subnuclear compartments, we measured FRAP recovery kinetics. Recovery of photo-bleached EGFP-RUNX2 proteins requires that non-bleached proteins migrate to the bleach zone, presumably while EGFP-RUNX2 proteins are in binding equilibrium with their cognate sites. In interphase cells RUNX2 is located in many small nuclear foci, which represent sites containing RNA polymerase II that are engaged in transcription or where transcription is poised for initiation (Young et al, 2007b;Javed et al, 2000;Zeng et al, 1998). After photo-bleaching, EGFP-RUNX2 recovery at these nuclear sites occurs with a half-time (t1/2) of 12 seconds (Fig. 2; see also Fig. 6 below). This finding is consistent with our previous results for RUNX2 in live cells (Harrington et al, 2002) and the general observation that recovery rates for most nuclear proteins are slow relative to diffusion (<500 milliseconds). The immobile fraction of RUNX2 is 35% reflecting a tightly bound fraction that does not exchange over the experimental time course (i.e., 1 to 2 minutes). Thus, RUNX2 has two populations at non-nucleolar foci, one exchanging rapidly and a second that is stably engaged in immobile complexes.
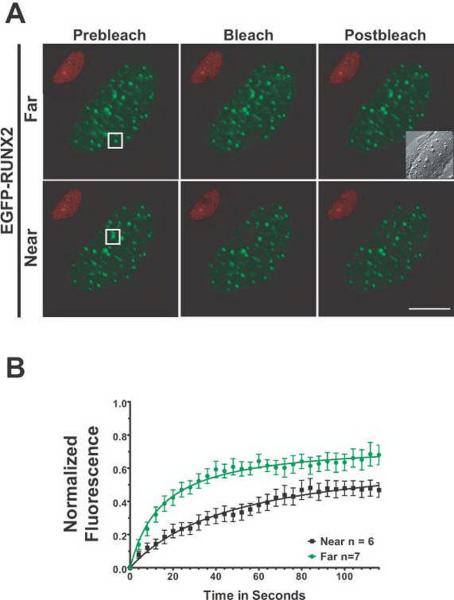
Fig. 6. RUNX2 has different binding kinetics at nucleolar versus subnuclear foci as measured by FRAP.
(A) The micrographs show the region of interest in cells before photobleaching, just after photobleaching, and 126 seconds later when the fluorescence had recovered. The size bar is 10 μm. (B) Normalized mean fluorescence recovery curves over time were calculated for RUNX2 foci at nucleoli and at subnuclear foci; the means were plotted with standard errors for each time point. Nucleolar RUNX2 foci recovered more slowly (29 seconds) and had a larger tightly bound immobile fraction (47%) than RUNX2 subnuclear foci (12 seconds, 35%). These results indicate a much stronger interaction of nucleolar RUNX2 with its binding site or partner proteins than the subnuclear RUNX2 foci.
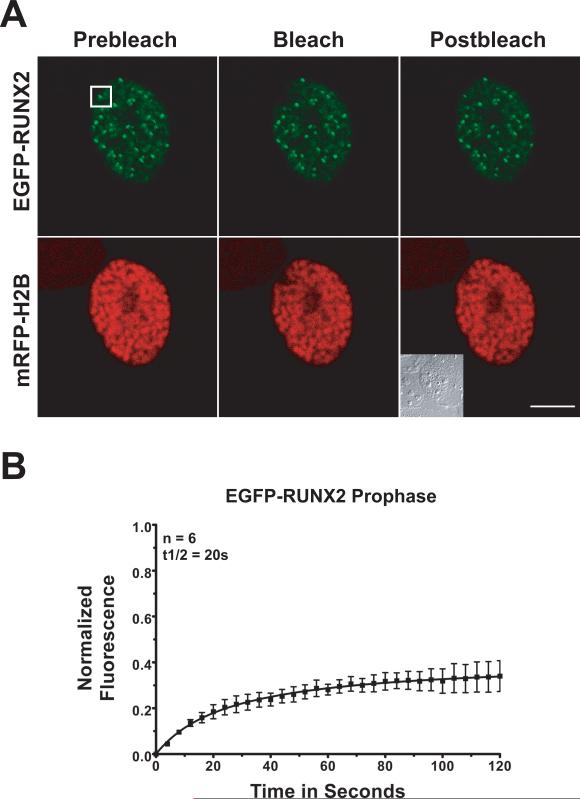
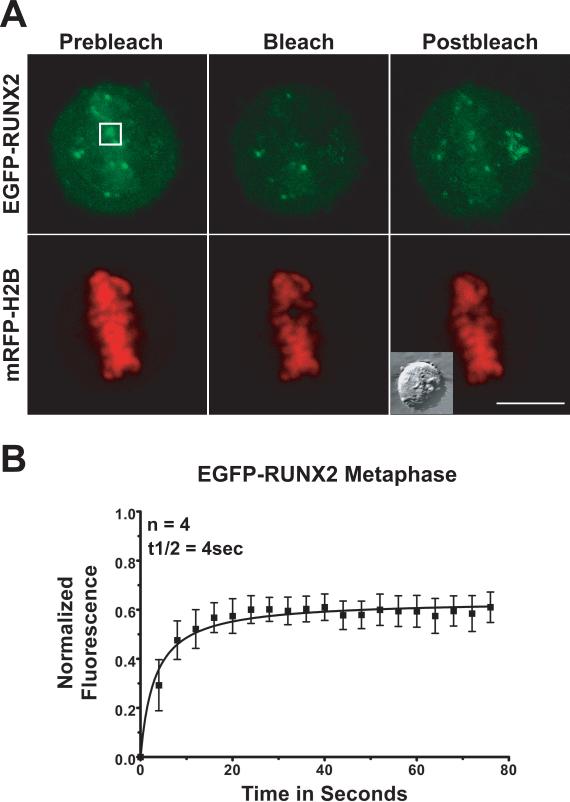
To evaluate changes in binding that may occur during mitosis, we photobleached EGFP-RUNX2 on condensed chromosomes at different mitotic stages (Table 1). During prophase (Fig. 3), photobleach recovery occurs with a half-time (t1/2) of ~20 seconds. This recovery rate is much slower than the initial recovery in the nuclear foci of interphase cells (12 seconds). The immobile RUNX2 fraction that is tightly bound is 67% at prophase, which is twice the amount of stably-bound RUNX2 in interphase foci. In contrast, in metaphase cells, RUNX2 exchange on chromosomes is much more rapid. After photobleaching, the exchanging population of EGFP-RUNX2 on metaphase chromosomes recovers with a t1/2 of ~4 seconds (Fig. 4). In metaphase the tightly bound non-exchanging immobile fraction is ~41%, a value closer to the ~35% fraction observed in interphase nuclear foci.
TABLE 1.
Kinetic Parameters of RUNX2
| t1/2 (seconds)* | Immobile Fraction (%) | n | |
|---|---|---|---|
| Interphase | |||
| Non-nucleolar | 12 | 35 ± 2.5 | 7 |
| Nucleolar | 29 | 47 ± 5.5 | 6 |
| Mitosis | |||
| Prophase | 20 | 67 ± 2.6 | 6 |
| Metaphase | 4 | 41 ± 3 | 4 |
| Telophase | 7 | 37 ± 2.5 | 6 |
t1/2 is the half time of photobleach recovery.
Fig. 3. Photobleach recovery of RUNX2 on chromosomes during prophase.
Multiple prophase cells expressing EGFP-RUNX2 and mRFP-Histone H2B were used for FRAP analysis. (A) The images show the region of interest in cells before photobleaching, just after photobleaching, and 120 seconds later when the fluorescence had recovered. The size bar is 10 μm. (B) Normalized mean fluorescence recovery curves over time for 6 cells were calculated for chromosome bound RUNX2; the means were plotted with standard errors for each time point. The kinetics of recovery measure the interaction of the protein with its binding site. Cells in prophase show a half-time of recovery (t1/2) of 20 seconds. The large immobile fraction (67%) represents RUNX2 in complexes that are tightly bound and do not dissociate over the time course of the experiment.
Fig. 4. Photobleach recovery of RUNX2 on chromosomes during metaphase.
Multiple metaphase cells expressing EGFP-RUNX2 and mRFP-Histone H2B were used for FRAP analysis. (A) The images show the region of interest in cells before photobleaching, just after photobleaching, and 80 seconds later when the fluorescence had recovered. The size bar is 10 μm. (B) Normalized mean fluorescence recovery curves over time were calculated for chromosome bound RUNX2; the means were plotted with standard errors for each time point. Cells in metaphase show a t½ of recovery of 4 seconds and the immobile fraction (41%) had decreased since prophase.
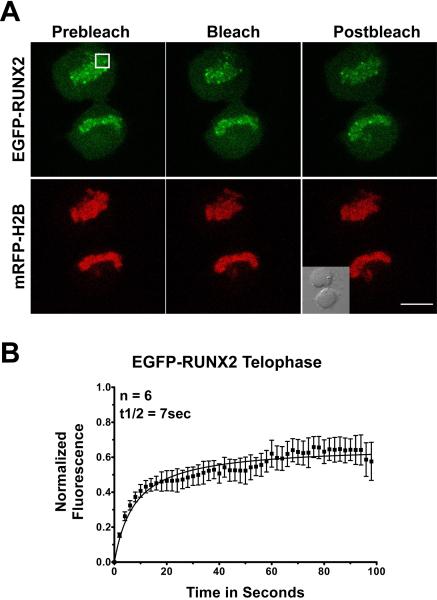
Chromosomes in anaphase cells are in very rapid motion, precluding reliable determination of photobleach recovery kinetics. When cells reach telophase, the exchanging pool of EGFP-RUNX2 recovers with a t1/2 of ~7 seconds (Fig. 5), similar to that in metaphase (~4 seconds) yet faster than in non-nucleolar foci during interphase (~12 seconds) (see below). The non-exchanging immobile fraction in telophase is 37%, similar to the values during metaphase and interphase. These results suggest that there are stage-specific changes in the binding interactions experienced by RUNX2 at chromatin as the cell progresses from interphase through mitosis.
Fig. 5. Photobleach recovery of RUNX2 on chromosomes during telophase.
Multiple telophase cells expressing EGFP-RUNX2 and mRFP-Histone H2B were used for FRAP analysis. (A) The micrographs show the region of interest in cells before photobleaching, just after photobleaching, and 100 seconds later when the fluorescence had recovered. The size bar is 10 μm. (B) Normalized mean fluorescence recovery curves over time were calculated for chromosome bound RUNX2; the means were plotted with standard errors for each time point. Cells in telophase had a t1/2 of recovery of 7 seconds and an immobile fraction of 37%.
The kinetics of RUNX2 binding are different at RNA polymerase I and RNA polymerase II target genes
RUNX2 is a transcriptional regulator of both protein coding genes in the non-nucleolar foci and rRNA genes at the nucleolus. To compare the binding properties of RUNX2 at these sites, we measured the photobleach recovery kinetics of EGFP-RUNX2 in the nucleus and adjacent to the nucleolus (Fig. 6). Recovery for the exchanging fraction at nucleolar-associated RUNX2 foci occurs with a t1/2 of ~29 seconds and the tightly bound immobile fraction is ~47%, reflecting a larger immobile population of RUNX2 molecules at nucleoli. Concurrently, RUNX2 exchanges more rapidly at non-nucleolar foci with a recovery half-time (t1/2), of ~12 seconds and an immobile fraction of ~35% (Figs. 6A and 6B). This is consistent with the existence of different binding interactions for RUNX2 at these two sites.
DISCUSSION
For the first time we have directly observed the mitotic choreography of epigenetic transcription factors in association with genes as live cells progress through cell division. Importantly, measurements of the binding affinities of these interactions by FRAP indicate that there are at least two mitotic populations of RUNX2: one which exchanges during the experimental time-course and the other immobile population which does not. Both the recovery half-time for the exchanging population and the immobile fraction of photo-bleached RUNX2 increase during prophase and subsequently decrease during telophase when chromatin begins to decondense in preparation for interphase. These changes in the kinetics and equilibrium of RUNX2 binding interactions are generally commensurate with the extent of chromosome condensation at different mitotic stages.
The increased chromosomal binding of RUNX2 during mitosis may relate to changes in protein/protein interactions with co-regulatory factors and/or post-translational modifications (e.g. phosphorylation and acetylation). For example, RUNX2 remains associated with its co-factor groucho/TLE1, but the histone deacetylase HDAC1 selectively dissociates from RUNX2 complexes during mitosis (Ali et al, 2010). In addition, RUNX2 is hyper-phosphorylated by CDK1/cyclin B during mitosis (Rajgopal et al, 2007) which may affect its binding affinity in multisubunit complexes linked to chromatin. Furthermore, our FRAP results suggest functional distinctions in RUNX2 association with RNA polymerase I and II promoters. Binding interactions of RUNX2 at nucleolar-associated sites, where RUNX2 may repress RNA polymerase I dependent rRNA genes, appear to be different from binding interactions of RUNX2 at subnuclear sites where RUNX2 regulates the expression of RNA polymerase II transcribed protein coding genes. Changes in protein conformation and promoter architecture that accompany global modifications in chromatin structure may also affect recovery rates and the immobile fraction during mitosis. A plausible scenario is that accessibility of binding sites for RUNX2 decreases as chromatin condenses. RUNX2 binding may be further affected by the discharge of co-regulatory proteins as chromatin at RUNX2-responsive genes transitions from transcriptionally active to silent at mitosis.
Transcriptional regulators such as RUNX2 bind to chromatin and to the nuclear matrix, recruit additional transcription factors and chromatin modifiers to promoters, and then engage the basal transcriptional machinery. Conventional in vitro techniques for studying transcription, such as chromatin immunoprecipitation and electrophoretic mobility shift assays, may leave the impression that binding of factors at promoters is entirely static. Advanced live cell techniques reveal a more dynamic binding of transcription factors at promoters. However, transcription factor dynamics have been studied using different species, different genes with different copy numbers, and with a variety of transcriptional regulators. The results have proven to be system specific.
The use of fluorescent fusion proteins expressed in cells containing chromosomally integrated arrays of tandemly repeated genes has shown that the binding of transcription factors, including the glucocorticoid receptor (McNally et al, 2000), the estrogen receptor (Sharp et al, 2006), the estrogen receptor coactivators CBP and SRC-1 (Stenoien et al, 2001), the progesterone receptor (Rayasam et al, 2005), and NF-kB (Bosisio et al, 2006), at target promoters can be highly dynamic. These transcription factors exchange over a time period of seconds, while over periods of 15 minutes to several hours some may cycle between being present on the promoter and being absent (discussed in (Carlberg and Seuter, 2010)). In the first example reported, the glucocorticoid receptor exchanged on a 200 copy array of the mouse mammary tumor virus promoter with a half-time of 5 seconds (McNally et al, 2000).
Studies of some native active genes in Drosophila using FRAP (Yao et al, 2006) or in yeast using biochemical assays (Nalley et al, 2006) have shown a more stable and constitutive binding of transcription factors. However, on one naturally occurring native array of CUP1 genes in yeast, the Ace1 transcription factor has a more dynamic exchange (~ 1 minute) as shown by FRAP and also cycles on and off the promoter over longer times (Karpova et al, 2008). This dependence of transcription factor kinetics on species, factor, promoter and copy number suggests that important questions remain to be answered about the role of factor exchange at promoters. The exchange of transcription factors on target genes as cells proceed through mitosis and the epigenetic consequences of that exchange have not been examined in any of these systems. Our experiments have addressed this deficiency by measuring the dynamic binding of RUNX2 to non-nucleolar compartments and nucleoli during interphase, as well as to condensing chromosomes during mitosis.
Our study has established dynamic parameters of epigenetic control by a transcription factor that is lineage-determining and cancer-related. Our findings that RUNX2 recovery rates and immobile fractions change transiently as cells enter and exit mitosis are consistent with a cellular mechanism that supports transcription factor retention during a cycle of chromatin reconfiguration (i.e., condensation followed by decondensation) and mitotic partitioning during cell division. A biological consequence of the modifications in retention of transcription factors on mitotic chromosomes is that phenotype-commitment or compromised control characteristic of cancer cells will persist during proliferative expansion.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rong-Lin Xie, Kaleem Zaidi and Akhter Ali for stimulating discussions. We also thank Charlene Baron for assistance with the presentation of digital images, as well as Judy Rask for assistance with manuscript preparation.
Contract Grant Sponsor: NIH Contract grant number: CA082834.
Footnotes
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Literature Cited
- Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, Shakoori AR, Montecino MA, Lian JB, van Wijnen AJ, Imbalzano AN, Stein GS, Stein JL. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci U S A. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SA, Zaidi SK, Dobson JR, Shakoori AR, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proc Natl Acad Sci U S A. 2010;107:4165–4169. doi: 10.1073/pnas.1000620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi R, Zaidi SK, Pande S, Hassan MQ, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The leukemogenic t(8;21) fusion protein AML1-ETO controls ribosomal RNA genes and associates with nucleaolar organizing regions at mitotic chromosomes. J Cell Sci. 2008;21:3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J, Mora-Bermudez F, Klee T, Daigle N, Ellenberg J. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys J. 2006;90:1878–1894. doi: 10.1529/biophysj.105.071241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwell JP, van Wijnen AJ, Fey EG, Dworetzky S, Penman S, Stein JL, Lian JB, Stein GS. Osteocalcin gene promoter-binding factors are tissue-specific nuclear matrix components. Proc Natl Acad Sci USA. 1993;90:3162–3166. doi: 10.1073/pnas.90.8.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlberg C, Seuter S. Dynamics of nuclear receptor target gene regulation. Chromosoma. 2010;119:479–484. doi: 10.1007/s00412-010-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero G, McDonald D, Crawford E, de VG, Hendzel MJ. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods. 2003;29:14–28. doi: 10.1016/s1046-2023(02)00288-8. [DOI] [PubMed] [Google Scholar]
- Choi J-Y, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein GS. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci, USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy CR, Xie R, Frederick D, Hussain S, Zaidi SK, Vradii D, Javed A, Li X, Jones SN, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Hum Mol Genet. 2010;19:1048–1057. doi: 10.1093/hmg/ddp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- Grummt I, Voit R. Linking rDNA transcription to the cellular energy supply. Cell Cycle. 2010;9:225–226. doi: 10.4161/cc.9.2.10614. [DOI] [PubMed] [Google Scholar]
- Harrington KS, Javed A, Drissi H, McNeil S, Lian JB, Stein JL, van Wijnen AJ, Wang YL, Stein GS. Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically associate with stationary subnuclear domains. J Cell Sci. 2002;115:4167–4176. doi: 10.1242/jcs.00095. [DOI] [PubMed] [Google Scholar]
- Jantzen HM, Admon A, Bell SP, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Javed A, Guo B, Hiebert S, Choi J-Y, Green J, Zhao S-C, Osborne MA, Stifani S, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Groucho/TLE/R-Esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113:2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, McNally JG. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010;339:189–195. doi: 10.1007/s00441-009-0832-8. [DOI] [PubMed] [Google Scholar]
- Kota KP, Wagner SR, Huerta E, Underwood JM, Nickerson JA. Binding of ATP to UAP56 is necessary for mRNA export. J Cell Sci. 2008;121:1526–1537. doi: 10.1242/jcs.021055. [DOI] [PubMed] [Google Scholar]
- Kruhlak MJ, Lever MA, Fischle W, Verdin E, Bazett-Jones DP, Hendzel MJ. Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J Cell Biol. 2000;150:41–51. doi: 10.1083/jcb.150.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele T, Oh P, Nickerson JA, Ingber DE. An improved mathematical approach for determination of molecular kinetics in living cells with FRAP. Mech Chem Biosyst. 2004;1:181–190. [PubMed] [Google Scholar]
- Lele T, Wagner SR, Nickerson JA, Ingber DE. Methods for measuring rates of protein binding to insoluble scaffolds in living cells: histone H1-chromatin interactions. J Cell Biochem. 2006;99:1334–1342. doi: 10.1002/jcb.20997. [DOI] [PubMed] [Google Scholar]
- Lele TP, Ingber DE. A mathematical model to determine molecular kinetic rate constants under non-steady state conditions using fluorescence recovery after photobleaching (FRAP) Biophys Chem. 2006;120:32–35. doi: 10.1016/j.bpc.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Merriman HL, van Wijnen AJ, Hiebert S, Bidwell JP, Fey E, Lian J, Stein J, Stein GS. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- Nalley K, Johnston SA, Kodadek T. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature. 2006;442:1054–1057. doi: 10.1038/nature05067. [DOI] [PubMed] [Google Scholar]
- Nickerson JA. The biochemistry of RNA metabolism studied in situ. RNA Biol. 2009;6:25–30. doi: 10.4161/rna.6.1.7563. [DOI] [PubMed] [Google Scholar]
- Pande S, Ali SA, Dowdy C, Zaidi SK, Ito K, Ito Y, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. J Cell Physiol. 2009;218:473–479. doi: 10.1002/jcp.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Gorski SA, Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 2004;375:393–414. doi: 10.1016/s0076-6879(03)75025-3. [DOI] [PubMed] [Google Scholar]
- Phair RD, Misteli T. Kinetic modelling approaches to in vivo imaging. Nat Rev Mol Cell Biol. 2001;2:898–907. doi: 10.1038/35103000. [DOI] [PubMed] [Google Scholar]
- Pikaard CS, Smith SD, Reeder RH, Rothblum L. rUBF, an RNA polymerase I transcription factor from rats, produces DNase I footprints identical to those produced by xUBF, its homolog from frogs. Mol Cell Biol. 1990;10:3810–3812. doi: 10.1128/mcb.10.7.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgopal A, Young DW, Mujeeb KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Mitotic control of RUNX2 phosphorylation by both CDK1/cyclin B kinase and PP1/PP2A phosphatase in osteoblastic cells. J Cell Biochem. 2007;100:1509–1517. doi: 10.1002/jcb.21137. [DOI] [PubMed] [Google Scholar]
- Rayasam GV, Elbi C, Walker DA, Wolford R, Fletcher TM, Edwards DP, Hager GL. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol Cell Biol. 2005;25:2406–2418. doi: 10.1128/MCB.25.6.2406-2418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TP, Ingber DE, Mancini MA. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J Cell Sci. 2006;119:4101–4116. doi: 10.1242/jcs.03161. Erratum in: J Cell Sci. 2006 Oct 15;119(Pt 20):4365. Lele, Tanmay T [corrected to Lele, Tanmay P] [DOI] [PubMed] [Google Scholar]
- Sprague BL, Muller F, Pego RL, Bungay PM, Stavreva DA, McNally JG. Analysis of binding at a single spatially localized cluster of binding sites by fluorescence recovery after photobleaching. Biophys J. 2006;91:1169–1191. doi: 10.1529/biophysj.105.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Nye AC, Mancini MG, Patel K, Dutertre M, O'Malley BW, Smith CL, Belmont AS, Mancini MA. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol Cell Biol. 2001;21:4404–4412. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Chiosea S, Ivshina M, Nickerson JA. In vitro FRAP reveals the ATP-dependent nuclear mobilization of the exon junction complex protein SRm160. J Cell Biol. 2004;164:843–850. doi: 10.1083/jcb.200307002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Chiosea S, Nickerson JA. The spatial targeting and nuclear matrix binding domains of SRm160. Proc Natl Acad Sci U S A. 2003;100:3269–3274. doi: 10.1073/pnas.0438055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang X-Q, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Choi J-Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, McNeil S, Pockwinse S, Nickerson JA, Shopland L, Lawrence JB, Penman S, Hiebert SW, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, van Wijnen AJ, Stein JL, Meyers S, Sun W, Shopland L, Lawrence JB, Penman S, Lian JB, Stein GS, Hiebert SW. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.