Abstract
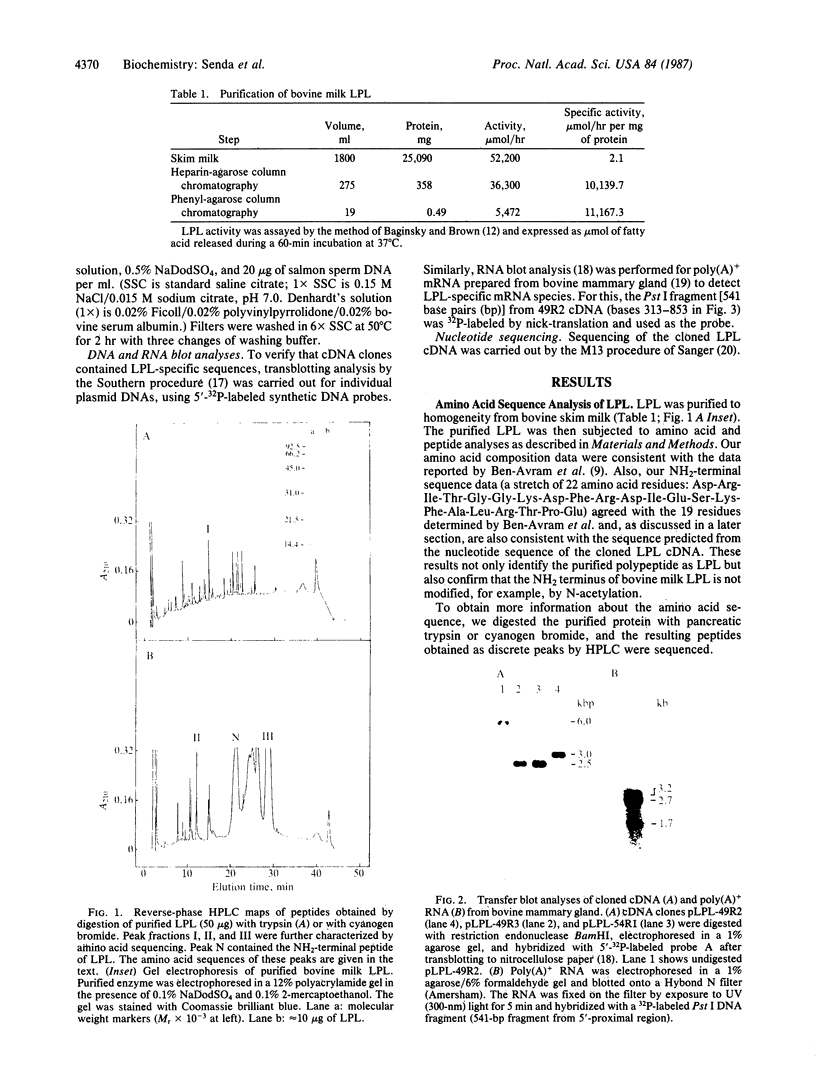
Lipoprotein lipase (LPL; triacylglycero-protein acylhydrolase, EC 3.1.1.34) was purified from bovine milk. Synthetic oligonucleotides were prepared, based on the amino acid sequences of three peptides obtained from partial digestion of purified LPL, and were used as probes to isolate cDNA clones for LPL mRNA from a bovine mammary gland. One of the clones, pLPL-49R2, contains an insert cDNA (49R2) of about 3.2 kilobases (kb) that hybridizes to all three probes and encodes a polypeptide that includes the NH2-terminal sequence of bovine LPL reported recently [Ben-Avram, C. M., Ben-Zeev, O., Lee, T. D., Hagga, K., Shively, J. E., Goers, J., Pedersen, M. E., Reeve, J. R. & Schotz, M. C. (1986) Proc. Natl. Acad. Sci. USA 83, 4185-4189]. Complete nucleotide sequence analysis revealed that cDNA insert 49R2 contains the entire coding region for LPL as well as a 3' untranslated region of about 1.6 kb. The predicted amino acid sequence indicates that bovine LPL is a hydrophilic protein consisting of 450 amino acids (Mr 50,548) in its unglycosylated form. Blot hybridization analysis of poly(A)+ mRNA from bovine mammary gland demonstrated that there are at least three sizes of LPL mRNAs--3.2, 2.5, and 1.7 kb--with the 2.5-kb mRNA being the most abundant. Restriction endonuclease mapping of other cDNA clones suggested that the variation in mRNA size results from differential utilization of polyadenylylation signals during mRNA processing.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky M. L., Brown W. V. A new method for the measurement of lipoprotein lipase in postheparin plasma using sodium dodecyl sulfate for the inactivation of hepatic triglyceride lipase. J Lipid Res. 1979 May;20(4):548–556. [PubMed] [Google Scholar]
- Beaumont J. L., Carlson L. A., Cooper G. R., Fejfar Z., Fredrickson D. S., Strasser T. Classification of hyperlipidaemias and hyperlipoproteinaemias. Bull World Health Organ. 1970;43(6):891–915. [PMC free article] [PubMed] [Google Scholar]
- Ben-Avram C. M., Ben-Zeev O., Lee T. D., Haaga K., Shively J. E., Goers J., Pedersen M. E., Reeve J. R., Jr, Schotz M. C. Homology of lipoprotein lipase to pancreatic lipase. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4185–4189. doi: 10.1073/pnas.83.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson G., Olivecrona T. Interaction of lipoprotein lipase with heparin-Sepharose. Evaluation of conditions for affinity binding. Biochem J. 1977 Oct 1;167(1):109–119. doi: 10.1042/bj1670109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cryer A. Tissue lipoprotein lipase activity and its action in lipoprotein metabolism. Int J Biochem. 1981;13(5):525–541. doi: 10.1016/0020-711x(81)90177-4. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidoni A., Benkouka F., De Caro J., Rovery M. Characterization of the serine reacting with diethyl p-nitrophenyl phosphate in porcine pancreatic lipase. Biochim Biophys Acta. 1981 Jul 24;660(1):148–150. doi: 10.1016/0005-2744(81)90120-0. [DOI] [PubMed] [Google Scholar]
- Hahn P. F. ABOLISHMENT OF ALIMENTARY LIPEMIA FOLLOWING INJECTION OF HEPARIN. Science. 1943 Jul 2;98(2531):19–20. doi: 10.1126/science.98.2531.19. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LaRosa J. C., Levy R. I., Herbert P., Lux S. E., Fredrickson D. S. A specific apoprotein activator for lipoprotein lipase. Biochem Biophys Res Commun. 1970 Oct 9;41(1):57–62. doi: 10.1016/0006-291x(70)90468-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mahoney J. R., Jr, Beutler B. A., Le Trang N., Vine W., Ikeda Y., Kawakami M., Cerami A. Lipopolysaccharide-treated RAW 264.7 cells produce a mediator that inhibits lipoprotein lipase in 3T3-L1 cells. J Immunol. 1985 Mar;134(3):1673–1675. [PubMed] [Google Scholar]
- Narimatsu H., Sinha S., Brew K., Okayama H., Qasba P. K. Cloning and sequencing of cDNA of bovine N-acetylglucosamine (beta 1-4)galactosyltransferase. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4720–4724. doi: 10.1073/pnas.83.13.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Nilsson-Ehle P., Garfinkel A. S., Schotz M. C. Lipolytic enzymes and plasma lipoprotein metabolism. Annu Rev Biochem. 1980;49:667–693. doi: 10.1146/annurev.bi.49.070180.003315. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983 Feb;3(2):280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. N., Maraganore J. M., Meredith S. C., Heinrikson R. L., Kézdy F. J. Isolation of an active-site peptide of lipoprotein lipase from bovine milk and determination of its amino acid sequence. J Biol Chem. 1986 Jul 25;261(21):9678–9683. [PubMed] [Google Scholar]
- Sanger F. Determination of nucleotide sequences in DNA. Science. 1981 Dec 11;214(4526):1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- Shimada K., Gill P. J., Silbert J. E., Douglas W. H., Fanburg B. L. Involvement of cell surface heparin sulfate in the binding of lipoprotein lipase to cultured bovine endothelial cells. J Clin Invest. 1981 Oct;68(4):995–1002. doi: 10.1172/JCI110354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]





