Figure 5.
The PB-I-Surface Localization of PDIL2;3 Depends on the Catalytic Active Sites.
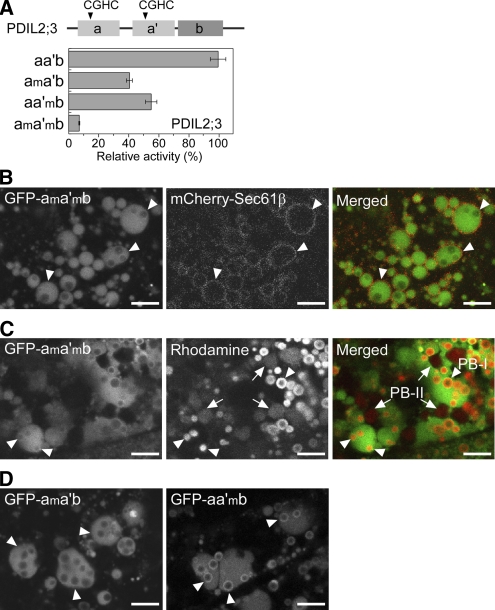
(A) Relative rRNase-refolding activities of recombinant PDIL2;3 variants. The arrowheads in the top panel indicate the positions of redox active sites of PDIL2;3: Cys59-X-X-Cys62 (a domain) and Cys195-X-X-Cys198 (a′ domain). The Cys residues in the active sites were substituted with Ala residues: Ala59-X-X-Ala62 (am domain) and Ala195-X-X-Ala198 (a′m domain). The wild type (aa′b) and three mutants (ama′b, aa′mb, and ama′mb) were purified, and their refolding activities were compared. The relative activities are presented as means with standard deviation of three replicates.
(B) Confocal fluorescence images of the endosperm cells (10 DAF) expressing both spGFP-PDIL2;3(ama′mb) and mCherry-Sec61β. The fluorescence signals of GFP-PDIL2;3(ama′mb) (left panel) and mCherry-Sec61β (middle panel) were converted to green and red, respectively, and merged (right panel). Arrowheads indicate PB-I in the ER lumen. Bars = 5 μm.
(C) PB-I (arrowheads) and PB-II (arrows) were visualized by Rhodamine staining. The fluorescence signals of GFP-PDIL2;3(ama′mb) (left panel) and Rhodamine (middle panel) were converted to green and red, respectively, and merged (right panel). Bars = 5 μm.
(D) Confocal fluorescence images of the endosperm (10 DAF) expressing spGFP-PDIL2;3(ama′b) (left panel) or spGFP-PDIL2;3(aa′mb) (right panel). The molecular masses of the fusion proteins were confirmed by immunoblot analysis. Arrowheads indicate PB-I in the ER lumen. Bars = 5 μm.