This work shows that the H3K4 trimethylase ATX1 regulates gene expression via mechanisms different from those reported for its yeast or mammalian homologs. It finds that ATX1 affects transcription at the promoters of target genes by a mechanism distinct from its ability to trimethylate lysine 4 of histone H3 within genes.
Abstract
The Arabidopsis thaliana trithorax-like protein, ATX1, shares common structural domains, has similar histone methyltransferase (HMT) activity, and belongs in the same phylogenetic subgroup as its animal counterparts. Most of our knowledge of the role of HMTs in trimethylating lysine 4 of histone H3 (H3K4me3) in transcriptional regulation comes from studies of yeast and mammalian homologs. Little is known about the mechanism by which ATX1, or any other HMT of plant origin, affects transcription. Here, we provide insights into how ATX1 influences transcription at regulated genes, playing two distinct roles. At promoters, ATX1 is required for TATA binding protein (TBP) and RNA Polymerase II (Pol II) recruitment. In a subsequent event, ATX1 is recruited by a phosphorylated form of Pol II to the +300-bp region of transcribed sequences, where it trimethylates nucleosomes. In support of this model, inhibition of phosphorylation of the C-terminal domain of Pol II reduced the amounts of H3K4me3 and ATX1 bound at the +300-nucleotide region. Importantly, these changes did not reduce the occupancy of ATX1, TBP, or Pol II at promoters. Our results indicate that ATX1 affects transcription at target genes by a mechanism distinct from its ability to trimethylate H3K4 within genes.
INTRODUCTION
Genetic, biochemical, and molecular characteristics of the ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1) have defined it as a plant counterpart of the Drosophila melanogaster trithorax (TRX) and mammalian mixed-lineage leukemia (MLL1) proteins (Alvarez-Venegas et al., 2003). MLL1, TRX, and ATX1 are members of multigene families related by the common SET (for suppressor of variegation, enhancer of zeste, and trithorax) domain, which carries a histone methyltransferase catalytic domain. ATX1, TRX, MLL1, and the related SET1 protein from the yeast Saccharomyces cerevisiae are capable of trimethylating lysine 4 of histone H3 (H3K4me3), a mark generally associated with transcriptionally active genes (Shukla et al., 2009). Phylogenetically, yeast SET1 segregates in a subgroup distinct from the larger, multidomain proteins in the ATX1, TRX, MLL1 (TRX) subgroup, reflecting the evolutionary divergence of the members of the two subgroups (Veerappan et al., 2008).
SET1 is the sole H3K4 methylase responsible for the global mono-, di-, and trimethyl H3K4 chromatin marks in yeast (Bernstein et al., 2002; Santos-Rosa et al., 2002). By contrast, the TRX subgroup members of animal and plant origin modify only a limited fraction of nucleosomes. MLL1 is required for the H3K4 trimethylation of <5% of promoters carrying this modification, including genes for developmental regulators, such as homeobox (Hox) genes (Milne et al., 2005), as well as genes involved in differentiation, organogenesis, leukemia, and stress responses (Wysocka et al., 2005; Ruthenburg et al., 2007). MLL1-regulated genes display lower levels of RNA Polymerase II (Pol II) and lower gene expression concomitant with the loss of H3K4me3 in MLL1–/– cells (Wang et al., 2009). The activity of ATX1 is similar to the animal MLL1 in this regard: ATX1 targets specific genes and is involved in maintaining normal levels of gene expression during development, transition to flowering, and organ identity, and regulates diverse classes of genes implicated in biotic and abiotic stress responses (Alvarez-Venegas and Avramova, 2001, 2005; Alvarez-Venegas et al., 2003; Pien et al., 2008; Saleh et al., 2008a, 2008b; Ding et al., 2009).
The genome-wide distribution of the H3K4me3 mark displays a predominantly gene-associated pattern with a strong bias toward the 5′-ends of actively transcribed genes, in yeast (Ng et al., 2003), animal (Wang et al., 2009), and plant genomes (Li et al., 2008; Zhang et al., 2009; van Dijk et al., 2010). The mechanisms determining H3K4me3 distribution, as well as its role in transcription, are still emerging (Kouzarides, 2007; Shilatifard, 2008; Cazzonelli et al., 2009; Shukla et al., 2009). The location and abundance of H3K4me3 marks appear to be affected by the transcriptional process, as Pol II transcription is required to recruit SET1 in yeast (Ng et al., 2003). A key feature distinguishing Pol II from RNA Polymerases I or III is the repetitive peptide sequence on the Pol II C-terminal domain (CTD). The CTD heptad consensus repeat [(Y1S2P3T4S5P6S7)n, where n ranges from 26 in yeast, 34 in Arabidopsis thaliana, to 52 in mammals] has different sites of phosphorylation within this repeat at different stages of transcription (Phatnani and Greenleaf, 2006). The phosphorylated forms of the CTD recruit different protein complexes to facilitate RNA processing (Fabrega et al., 2003) and chromatin modification (Ng et al., 2003).
The main stages of the transcription process are associated with distinct phosphorylation states of the CTD. Specifically, the formation of the preinitiation complex (PIC), transcription initiation, and the elongation and polyadenylation/termination stage each are associated with a distinct pattern of CTD phosphorylation. During the formation of PIC on the promoter DNA/chromatin template, transcription activators bind the coactivator protein complex, Mediator (Lee et al., 1999), which binds to the predominantly nonphosphorylated or hypophosphorylated form of Pol II and class II basal transcription factors (TFII). The TFIID component of PIC contains the TATA binding protein (TBP) that binds the TATA box sequence of promoters, as well as other TBP-associated factors (TAFs) or TBP-related factors involved in binding to promoters, particularly in those lacking a TATA box (Müller et al., 2007; Juven-Gershon and Kadonaga, 2010). Once incorporated into PIC, Mediator stimulates the cyclin-dependent kinase 7 (CDK7) component of TFIIH, which phosphorylates serine 5 (Ser5P) and serine 7 of the CTD heptad repeat. The Ser5P modification helps release Pol II from the Mediator/TFIID/TFIIA/TFIIH/TFIIE PIC complex, allowing Pol II to escape the promoter and to initiate transcription. Pol II retains its Ser5P modification predominantly during the transcription of the first several hundred nucleotides. Therefore, Ser5P is found primarily at the promoter regions and 5′-ends of genes and is considered a biochemical marker for transcription initiation and early elongation (Gomes et al., 2006; Chapman et al., 2007; Kim et al., 2009).
Further transcript elongation is associated with increased phosphorylation of serine 2 (Ser2P), initially occurring as a Ser2P/Ser5P form of the CTD repeat, and later transitioning to a Ser2P form (Komarnitsky et al., 2000; Egloff and Murphy, 2008). Ser2P is mediated by CDK9, which is recruited via a Ser5P-dependent mechanism (Qiu et al., 2009). The Ser2P modification recruits factors for mRNA polyadenylation and termination (McCracken et al., 1997; Birse et al., 1998). Thus, the Ser5P and Ser2P modifications occur after PIC formation and facilitate transcription initiation/elongation and recruitment of proteins involved in RNA processing and chromatin modification.
In yeast, recruitment of the H3K4 methylating SET1/COMPASS (for Complex Proteins Associated with SET1) complex to target genes requires prior initiation of transcription as it depends on the binding of SET1/COMPASS to the Ser5P form of the CTD (Ng et al., 2003). Thereby in yeast, the CTD phosphorylation state is involved in the positioning of the H3K4me3 mark. By contrast, the order of events in mammalian cells is apparently different, as the presence of H3K4me3 facilitates transcription initiation on MLL1-dependent promoters. Mammalian TFIID binds H3K4me3 through the plant homeodomain (PHD) finger of its TAF3 subunit, which serves to anchor TFIID to H3K4 trimethylated nucleosomes on activated MLL1-dependent promoters (Vermeulen et al., 2007). Thus, presence of H3K4me3 is necessary for efficient transcription from MLL1-dependent genes (Wang et al., 2009), illustrating the divergent roles of the SET1- and TRX-type histone methyltransferases and the order of appearance of H3K4me3 marks relative to transcription initiation.
Little is known about the mechanism by which ATX1, or any other TRX-related methyltransferase of plant origin, affects transcription. Here, we examined the role of ATX1 in regulating the transcription of three genes that are strongly affected by ATX1 in vegetative tissues. WRKY DNA BINDING PROTEIN70 (WRKY70), a member of the WRKY family of transcription factors, is involved in salicylic acid and jasmonic acid signaling pathways, and its transcription is regulated in part by ATX1 binding (Alvarez-Venegas et al., 2007). The LIPID TRANSFER PROTEIN7 (LTP7) gene, a member of an antimicrobial peptide family (García-Olmedo et al., 1998), is highly expressed in leaves and its expression is regulated by ATX1 binding (Alvarez-Venegas and Avramova, 2005). The 9-CIS-EPOXYCAROTENOID DIOXYGENASE (NCED3) gene encodes the rate-limiting enzyme in abscisic acid biosynthesis (Qin and Zeevaart, 1999) and is regulated by ATX1, as described here. Our studies demonstrate that ATX1’s role in Pol II recruitment is distinct from its role in H3K4 trimethylation.
RESULTS
Distribution Profiles of ATX1, Ser2P, Ser5P, and H3K4me3
As a first step toward understanding the interplay between ATX1, H3K4me3, and Pol II during transcription, we measured the distribution profiles of ATX1, H3K4me3, and the two main CTD phosphorylated forms of Pol II in ATX1 and atx1 genotypes (the atx1-1 mutant allele in our initial study [Alvarez-Venegas et al., 2003] is referred to here as atx1). Ser5P is indicative of the transition from PIC formation in the promoter region to transcription initiation and of early transcription elongation at the 5′ gene regions (Buratowski, 2009). The Ser2P form is indicative of later stages of elongation and termination (Komarnitsky et al., 2000; Egloff and Murphy, 2008). ATX1, H3K4me3, and the CTD modifications were measured by chromatin immunoprecipitation (ChIP), followed by quantitative PCR analysis of the amount of DNA enrichment for three ATX1-regulated genes (Figure 1A). ACTIN7 (ACT7) is not regulated by ATX1 and served as an ATX1-independent internal control in each analysis. The amount of DNA enrichment was measured at multiple points along the three ATX1-regulated genes by quantitative PCR (Figures 1B to 1E).
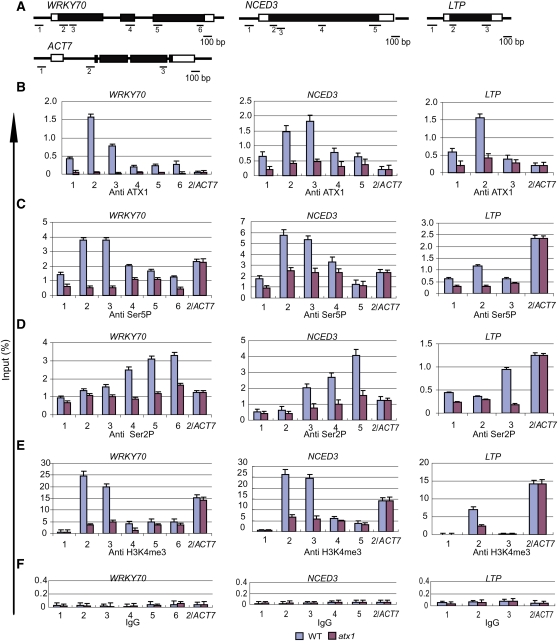
Figure 1.
Distribution of ATX1, Ser5P, and Ser2P of the CTD of Pol II and H3K4me3 on ATX1-Regulated Genes.
(A) Schematic diagrams of the WRKY70, NCED3, LTP, and ACT7 genes. Within the transcribed region of each gene, the 5′ or 3′ untranslated regions are shown as open boxes, the exons as black boxes, and the introns as thin black lines. The locations of the gene regions analyzed by ChIP-PCR are shown below each gene, and the corresponding sequences are in Supplemental Table 2 online. Region 1 is in the promoter region of each gene.
(B) to (F) The amounts of ATX1, the phosphorylated forms (Ser5P or Ser2P) of the CTD of Pol II, H3K4me3, or nonspecific binding of control IgG serum (IgG) at different regions of the genes were determined by ChIP-PCR. The gene regions analyzed by quantitative PCR are indicated on the x axis, and the DNA enrichment on the y axis is relative to the input DNA. ACT7 is not regulated by ATX1. The region analyzed for this gene corresponds to region 2 (2/ACT7), and data from the analyses were included in the profiles for each gene as an internal control. Experiments were repeated at least three times, each with three replicates, and the representative experiments shown indicate the mean + se, n = 3 replicates. WT, wild type.
[See online article for color version of this figure.]
The ATX1, Ser5P, Ser2P, and H3K4me3 levels were strongly affected by the atx1 mutation for WRKY70, NCED3, and LTP but not for the internal control ACT7 (Figures 1B to 1F). The results indicate that a functional ATX1 is required for the abundance of both forms of phosphorylated Pol II (Figures 1C and 1D) and of the H3K4me3 marks (Figure 1E) at the ATX1-regulated genes but not at the ATX1-independent ACT7. The large decreases in the Pol II Ser5P (Figure 1C) and Ser2P levels in atx1 mutants (Figure 1D) suggested that ATX1 regulated both the early and late stages of transcription (see further below).
The genotype-dependent difference in the recovery of ATX1-linked DNA provided evidence that the majority of the signal in the wild-type genotype was due to bound ATX1 (Figure 1B). In addition to the main peak at the +300-bp region (corresponding to region 2 in Figure 1A), the amount of ATX1 was somewhat higher across the gene, including the promoter region (corresponding to region 1 in Figure 1A). This suggested that ATX1 was present at the promoters of regulated genes. Alternatively, the peak at the +300-bp region could cause the signal at the adjacent region 1 via DNA linkage of these regions. To resolve these alternatives, we analyzed the H3K4me3 profiles.
The distribution of the H3K4me3 marks (Figure 1E) was similar to the ATX1 distribution profiles, except at the promoter regions (region 1), which contained ATX1 but not H3K4me3 marks. The lack of H3K4me3 enrichment in region 1 indicated this region was not affected by the signal peak at the adjacent region (+300 bp, region 2). This result argues that the ATX1 signals measured in the promoter regions are not due to DNA linkage to the adjacent +300-bp region (region 2). Therefore, the presence of ATX1 in the promoter regions occurred in a region lacking H3K4me3. This unexpected observation was investigated further below.
Biochemical models indicate that Ser5P occurs after the initiation of transcription and Ser2P occurs after the phosphorylation of Ser5 (Buratowski, 2009). The rapid addition of Ser5P often results in the detection of this modification in promoter regions (Gomes et al., 2006; Chapman et al., 2007; Kim et al., 2009; Qiu et al., 2009), but the detection of Ser2P at the promoter region of the genes was unusual. To examine whether the signals might originate from lack of specificity of the antibodies, we investigated the specificities of both the Ser5P and Ser2P antibodies against synthetic peptides. The synthetic peptides contained four units of the CTD consensus heptad repeat that were either nonphosphorylated or contained Ser2P or Ser5P modifications. As shown in Supplemental Figure 1 online, the Ser2P antibody recognized the nonphosphorylated form of CTD at ~10% of the efficiency of recognizing Ser2P, while the Ser5P antibody was specific for the Ser5P modification. These results indicated that the Ser5P distribution profile was accurate, but the Ser2P profile probably contained contributions from the nonphosphorylated form of CTD, accounting for the signal observed in the promoter regions.
ATX1 Binds the CTD of RNA Pol II
The similarity in the distribution profiles of Ser5P and ATX1 on ATX1 target genes suggested that an interaction between Pol II and ATX1 might be occurring. To test this possibility, the interactions of the ATX1 protein, as well as fragments of it (Figure 2A), with the CTD of Pol II were analyzed by yeast two-hybrid assays. The intact ATX1 protein bound strongly to the CTD (Figure 2B). Within ATX1, the ATX N-terminal fragment (ATX1N) did not bind but the ATX1 C-terminal fragment (ATX1C) did. Detailed analyses of ATX1C regions indicated that the ATX1DH fragment containing the DAST (for Domain Associated with SET in Trithorax, also referred to as FYRN-FYRC) and ePHD (for extended plant homeodomain) domains did not interact, but the ATX1 SET domain did interact with the CTD domain (Figure 2B). No interactions were observed when the DNA binding domain alone was used as bait for any of the ATX1 protein domains tested (Figure 2B).
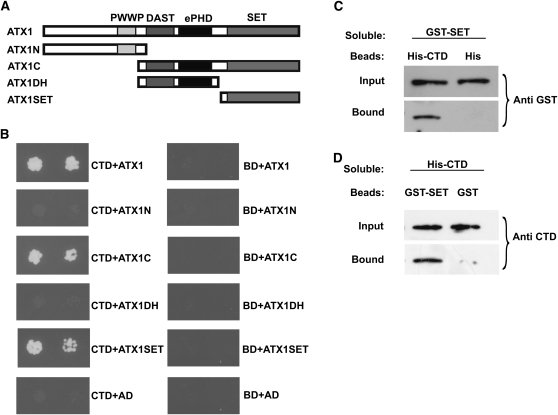
Figure 2.
ATX1 Interacts with the CTD of Pol II.
Yeast two-hybrid or in vitro interaction assays were performed.
(A) The different regions of ATX1 containing the indicated domains (PWWP, DAST, ePHD, and SET) that were tested in yeast two-hybrid analyses are shown.
(B) The DNA binding domain (BD) or BD-CTD fusions (CTD) were tested for binding to the activation domain (AD) fused to the various ATX1 domains shown in (A). The growth of two dilutions (4 × 10−3 and 8 × 10−4) of the yeast culture on SD medium lacking Trp, Leu, His, and adenine is shown.
(C) and (D) Representative immunoblots of the input amounts of a soluble protein or the amount of this protein bound to beads containing a surface-bound control tag or a tag fusion protein. The soluble protein being tested is denoted at the top of each panel.
(C) Beads containing a His tag (His) or a His tag fused to the CTD of Pol II (His-CTD) were assayed for their ability to bind a soluble GST fusion to the SET domain of ATX1 (GST-SET). Input or bound protein was detected with antibody to GST (Anti GST).
(D) Beads containing a GST tag or a GST tag fused to the SET domain of ATX1 (GST-SET) were assayed for their ability to bind a soluble CTD of Pol II fused to a His tag (His-CTD) and detected with antibody to the CTD of Pol II (Anti CTD). All experiments were repeated three times.
The yeast two-hybrid interactions were verified by in vitro pull-down assays. A protein containing glutathione S-transferase (GST) fused to the SET domain of ATX1 (GST-SET) was observed to bind to beads containing a His fusion to the CTD of Pol II but not to beads containing the His tag alone (Figure 2C). In the complementary experiment, bead-attached GST-SET, but not the GST control, was observed to bind the soluble His-tagged CTD of Pol II (Figure 2D), providing further evidence that the ATX1 SET domain binds directly to the CTD of Pol II.
ATX1 Preferentially Binds the Ser5P Form of the CTD of Pol II
The above assays could not distinguish whether the ATX1 SET domain showed a preference for the nonphosphorylated or a particular phosphorylated form of the CTD consensus repeat. This question was addressed by testing nonphosphorylated or phosphorylated forms of CTD peptides for their ability to bind to the SET domain of ATX1 (Figure 3A). This GST-SET protein bound to the nonphosphorylated form of the CTD peptide, but the strongest interaction was displayed when the CTD peptide contained Ser5P, and binding to the Ser2P form was not detectable (Figure 3A).
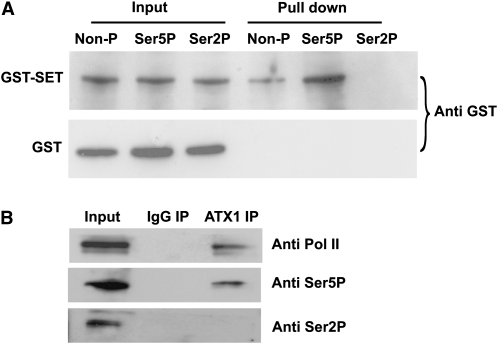
Figure 3.
Binding of the ATX1-SET Domain or Endogenous ATX1 to Different Phosphorylated Forms of the CTD of Pol II.
(A) Binding of soluble GST-SET or GST to bead-bound peptides containing four consensus CTD heptad repeats [(Y1S2P3T4S5P6S7)4] was measured. The 28mer peptides were either nonphosphorylated (Non-P), phosphorylated at Ser5P, or phosphorylated at Ser2P. The amount of GST or GST-SET protein bound to the peptides on the beads was determined by immunoblot analysis with antibody to GST (Anti GST).
(B) Representative immunoblots of the proteins immunoprecipitated with nonimmune IgG serum (IgG IP) or antibody to ATX1 (ATX1 IP) and detected with antibodies to total Pol II (Anti Pol II), the Ser5P form of the CTD of Pol II (Anti Ser5P), or the Ser2P form of the CTD of Pol II (Anti Ser2P) are shown. All experiments were repeated at least three times.
Next, these interactions were confirmed in vivo. Nuclear extracts were immunoprecipitated with anti-ATX1 antibodies and analyzed for the presence of the specifically phosphorylated forms of the CTD of Pol II. Total Pol II (independent of phosphorylation state) and the Ser5P form of Pol II were detected in the ATX1-immunoprecipitated sample, but the Ser2P form was not (Figure 3B). These data support a protein–protein interaction between ATX1 and the nonphosphorylated Pol II as well as between ATX1 and Ser5P of Pol II. The similar profiles of ATX1 and the Ser5P form of Pol II at the 5′-ends of these genes likely reflect these interactions. However, the occurrence of ATX1 at the promoter regions results from a different interaction, as described below.
ATX1 Affects TBP and Pol II Recruitment to Promoter Regions
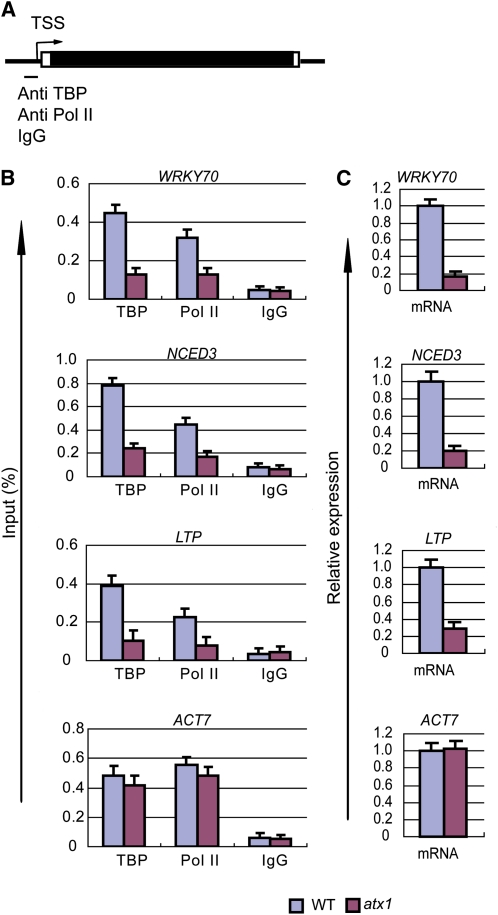
To analyze ATX1 effects on Pol II recruitment, we determined TBP and Pol II occupancy at the promoter regions in ATX1 or atx1 genotypes by ChIP-PCR with antibodies that recognize TBP or total Pol II (Figure 4). TBP was detected with a commercially available antibody against mammalian TBP that recognized a highly conserved region in Arabidopsis TBP1 and TBP2 (see Supplemental Figure 2 online). There was a large reduction of TBP and Pol II occupancy at the WRKY70, NCED3, and LTP promoters in atx1 relative to the ATX1 genotype (Figure 4B). These reductions in TBP and Pol II levels were associated with comparable reductions in the mRNA levels from WRKY70, NCED3, and LTP (Figure 4C). We note that the TBP and Pol II levels at the ACT7 promoter and ACT7 transcript levels were not affected by genotype (Figures 4B and 4C), consistent with the ATX1-independent transcription of ACT7. These results indicate that ATX1 affects transcription at ATX1-regulated genes by influencing the levels of resident TBP and Pol II proteins.
Figure 4.
ATX1 Affects the Amounts of TBP and Pol II Bound to Promoters and the Amount of mRNA Produced from ATX1-Regulated Genes.
ChIP-PCR of the promoter regions of the indicated genes was performed with antibodies to TBP or the N-terminal region of Pol II (recognition of this region is independent of phosphorylation in the CTD region), or mRNA levels were measured, in wild-type or atx1 backgrounds.
(A) A general gene representation indicating the promoter region and transcription start site (TSS) in the genes analyzed by ChIP-PCR with the indicated antibodies or nonimmune IgG (IgG). For each gene, the promoter region corresponds to region 1 in Figure 1A.
(B) The amounts of DNA detected by ChIP-PCR analysis for TBP, total Pol II, or nonimmune IgG bound to the promoter regions are shown.
(C) The relative transcript levels were determined in wild-type (WT) or atx1 rosettes of 20-d-old plant genotypes. All experiments were repeated at least three times. The bars represent the mean + se for representative experiments, n = 3 replicates.
[See online article for color version of this figure.]
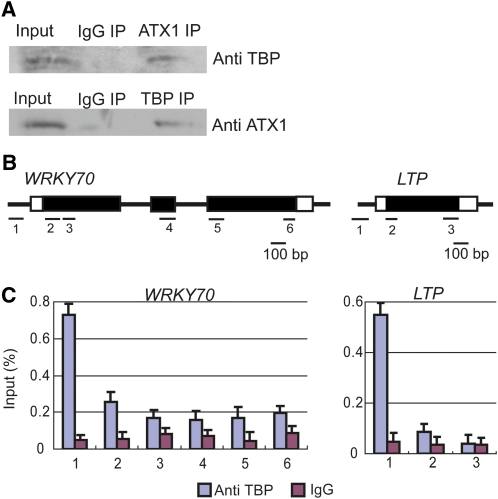
ATX1 and TBP Are Present in a Protein Complex
The occurrence of ATX1 and TBP at the promoter regions of ATX1-regulated genes and the dependence of the TBP level on ATX1 suggested a possible interaction between the two. To test this, we performed coimmunoprecipitation experiments. Nuclear extracts immunoprecipitated with antibody to ATX1 contained TBP (Figure 5A). Reciprocally, samples immunoprecipitated with antibody to TBP contained ATX1 (Figure 5A).
Figure 5.
ATX1 and TBP Interact in Vivo, and TBP Is Localized to Promoters.
(A) Nuclear extracts were immunoprecipitated with nonimmune IgG (IgG IP), ATX1 (ATX IP), or TBP antibodies (TBP IP). Immunoblots of the proteins immunoprecipitated by ATX1 antibody were analyzed with antibody to TBP (Anti TBP), while those immunoprecipitated by TBP antibody were analyzed with antibody to ATX1 (Anti ATX1). Representative immunoblots from three independent experiments are shown.
(B) A diagram of the gene regions analyzed by ChIP-PCR with TBP antibody or nonimmune IgG (see Supplemental Table 2 online for primer sequences).
(C) TBP or nonimmune IgG profiles of WRKY70 and LTP. The amount of DNA detected by ChIP-PCR, as a percentage of input DNA (y axis), is shown for each region. Experiments were repeated twice, and the bars represent the mean + se of a representative experiment, n = 3 replicates.
[See online article for color version of this figure.]
A possible experimental artifact leading to the observed interaction between ATX1 and TBP could potentially occur through a DNA linkage spanning separate ATX1 and TBP locations, although this linkage would be expected to be limited as these samples were not chemically cross-linked. We tested for a DNA-dependent linkage of these proteins as follows. First, we confirmed that TBP is localized only on promoters (Figures 5B and 5C). Next, a coimmunoprecipitation experiment was performed in the presence or absence of DNase I. DNase I treatment reduced DNA levels of target promoters by 200- to 1000-fold without reducing the amount of coprecipitated ATX1 and TBP (see Supplemental Figure 3 online). Additionally, the immunoprecipitated proteins appeared to be free of DNA, as very low levels of DNA were recovered in the immunoprecipitated fraction (see Supplemental Figure 3 online). These results indicate that the coimmunoprecipitation of ATX1 and TBP was not DNA dependent. Therefore, in light of TBP’s localization to promoter regions (Figure 5C), we conclude that the occurrence of ATX1 within a protein complex containing TBP supports a role for ATX1 at ATX1-dependent promoters.
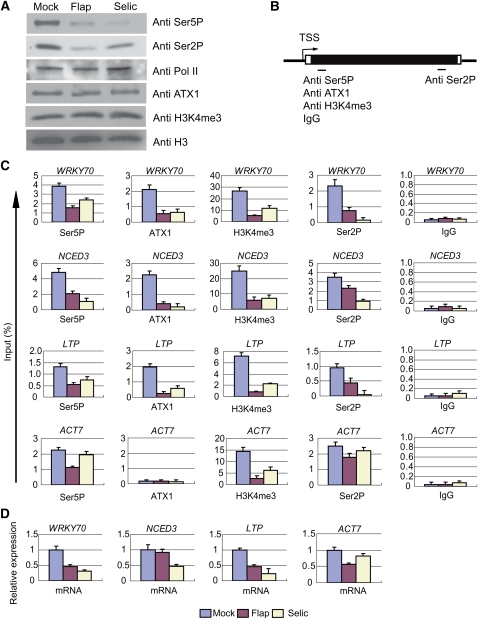
Inhibition of Ser5 Phosphorylation in Arabidopsis
Our results above showed that ATX1 interacted with the Ser5P form of Pol II. The concurrent changes in the amounts of ATX1, Ser2P, Ser5P, and H3K4me3 in the atx1 background, however, complicated any mechanistic interpretation. We sought to reduce the complexity of the analysis by altering Ser5P levels in an ATX1 genotype. As Arabidopsis kinases specific for the phosphorylation of Ser2 or Ser5 are not clearly established (Shimotohno et al., 2003; Umeda et al., 2005), we analyzed the effectiveness of selective kinase inhibitors as an alternative method of reducing Ser5P levels. Flavopiridol (Flap) or Seliciclib (Selic) can inhibit CDK7 and CDK9 in mammalian cells, which are predominantly responsible for phosphorylation of Ser5 and Ser2, respectively (Shapiro, 2006). Each drug diminished overall Ser2P and Ser5P levels in Arabidopsis (Figure 6A), consistent with the ability of either of these drugs to inhibit both CDK7 and CDK9 (Shapiro, 2006). The total levels of Pol II, ATX1, H3K4me3, or H3 were unchanged (Figure 6A).
Figure 6.
Chemical Inhibition of CDK7/CDK9-Like Kinases Reduces ATX1, Ser5P, Ser2P, and H3K4me3 Levels within Target Genes.
Mock-, Flavopiridol (Flap)-, or Seliciclib (Selic)-treated leaves were analyzed after 6 h of treatments.
(A) The amount of specific proteins or protein modifications in nuclear extracts of mock- or inhibitor-treated samples was determined with the indicated antibodies.
(B) A general representation of the gene regions analyzed by ChIP-PCR for the indicated antibodies or nonimmune IgG (IgG). The region analyzed for Ser5P, ATX1, H3K4me3, or IgG corresponds to region 2 for each gene, and the region analyzed for Ser2P corresponds to region 6 for WRKY70, region 5 for NCED3, or region 3 for LTP and ACT7 (see Figure 1A and/or Supplemental Table 2 online).
(C) The genes analyzed by ChIP-PCR are shown above each panel with the antibody or IgG designated at the bottom of each panel. The treatment color key is at the bottom of the figure.
(D) The levels of mRNA relative to rRNA were determined. rRNA was chosen as an internal control because rRNA genes are transcribed by RNA Polymerase I, which lacks a CTD and therefore should not be affected by the Flap or Selic inhibitors. Experiments were repeated three times. The bars represent the mean + se of a representative experiment, n = 3 replicates.
Effects of Ser5 Phosphorylation on ATX1 Recruitment at the +300-bp Region
Treatment with either Flap or Selic reduced both Ser2P and Ser5P Pol II forms at the three ATX1 regulated genes and at the ATX1-independent ACT7 gene as well (Figures 6B and 6C). It is important to note that the relative H3K4me3 levels were also considerably lower at the +300-bp region of all four genes. By contrast, the relative levels of ATX1 were strongly reduced at the ATX1-dependent WRKY70, NCED3, and LTP genes, but there was no change in the background ATX1 levels at the ATX1-independent ACT7 gene. The results indicate that inhibition of Ser2P/Ser5P levels resulted in reduced H3K4me3 levels at both ATX1-dependent and -independent genes. This suggests that the different histone methyl transferase(s) involved in trimethylating H3K4 at ACT7 also require Ser2P or Ser5P for their recruitment and/or activity. We conclude that Ser5P and/or Ser2P were required for normal levels of ATX1 and H3K4me3 at the +300-bp region of ATX1-regulated genes.
Uncoupling H3K4me3 from mRNA Levels
In the presence of Flap or Selic, the transcript levels produced from the three ATX1-target genes were decreased to 50% or less of control mRNA levels (Figure 6D). One exception was NCED3 mRNA in the Flap-treated sample, as the mRNA levels were comparable to those in the mock control (Figure 6D). This result demonstrates that the NCED3 gene can efficiently produce transcripts despite the reduced levels of Ser5P, H3K4me3, and ATX1 at the +300-bp region. This suggests that NCED3 is less dependent on the Ser5P and H3K4me3 modifications than are WRKY70 and LTP. The ACT7 transcripts were also only slightly reduced in the presence of Selic, despite the strong reduction in H3K4me3 levels (Figure 6D). These results are intriguing because they illustrate that efficient transcription and/or transcript processing could be uncoupled from the levels of Ser5P and H3K4me3 for two of the four genes examined. By contrast, the transcript levels from all tested genes, including ACT7, correlated with the levels of Ser2P (Figures 6C and 6D).
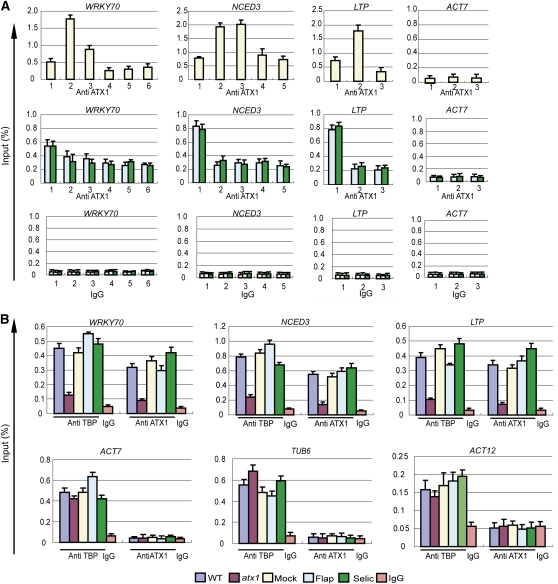
Uncoupling ATX1 Binding at Promoters and at Transcribed Regions
Our observation that Flap or Selic treatments resulted in diminished ATX1 signals at the +300-bp regions (Figure 6C) provided an opportunity to examine whether ATX1 binding to promoters occurred during these conditions. For a better basis for comparison, we analyzed the distribution of ATX1 along the length of the WRKY70, NCED3, LTP, and ACT7 genes (Figure 7A). In the mock-treated samples, the ATX1 distribution at the ATX1-regulated genes displayed the pattern seen in nontreated wild-type cells, with the characteristic peak at +300 bp (Figure 7A, top row). The ATX1 profile at the nontarget ACT7 gene had the expected pattern of low ATX1 amounts (Figure 7A, top row). However, treatment with either Flap or Selic dramatically changed the ATX1 profiles of the ATX1-regulated genes. These genes had diminished levels of ATX1 within the transcribed regions but retained ATX1 at their promoter regions (Figure 7A, middle row). By contrast, the profile of the ACT7 gene did not change (Figure 7A, middle row). This result demonstrates that ATX1 promoter occupancy was independent of ATX1 accumulation within the transcribed region. Most importantly, under these conditions, the undiminished ATX1 signal at the promoters was the strongest signal in the profiles of the ATX1-targeted genes (Figure 7A, middle row). Therefore, the ability of the inhibitors to diminish ATX1 signals in the +300-bp regions without reducing the amount of ATX1 occupancy at the promoters clearly demonstrates that the peak of ATX1 at the promoters was not a technical artifact derived from the adjacent +300-bp region signal. We conclude that ATX1 promoter occupancy was independent of ATX1 accumulation within the transcribed region.
Figure 7.
Inhibitors of CDK7/CDK9-Like Kinases Diminish ATX1 Binding within Genes but Do Not Lower ATX1 or TBP Levels at Promoters.
The effects of genotype (wild type (WT) or atx1) or 6 h of treatment with mock or CDK7/CDK9 inhibitors (mock, Flap, or Selic) were analyzed by ChIP-PCR for changes in TBP or ATX1 occupancy on selected gene regions in leaves. The color key for the genotype or treatment is at the bottom of the figure. The gene name is shown above each panel, and the antibody or nonimmune IgG serum (IgG) used for ChIP-PCR is designated below the lanes.
(A) The effects of mock, Flap, or Selic treatments on ATX1 profiles on the WRKY70, NCED3, LTP, and ACT7 genes. The numbers on the x axis show the gene regions analyzed and correspond to the regions diagrammed in Figure 1A. Region 1 corresponds to the promoter region of each gene.
(B) The occupancy of TBP and ATX1 at the promoter region (region 1 in Figure 1A and/or in Supplemental Table 2 online) of three ATX1-regulated genes (WRKY70, NCED3, and LTP) and three genes not regulated by ATX1 (ACT7, TUB6, and ACT12). Experiments (A) were performed twice, and experiments in (B) were performed three times for WRKY70, NCED3, and LTP and twice for ACT7, TUB6, and ACT12. The bars represent the mean + se of a representative experiment, n = 3 replicates.
ATX1 Occupancy at Promoters Is Required for TBP Binding to Promoters
As observed above, TBP promoter occupancy was reduced in an atx1 genotype (Figure 4), but the nature of this dependency on ATX1 was unclear. The ability of Flap and Selic treatments to reduce ATX1 levels within transcribed regions, but not at promoter regions (Figure 7A), provided an opportunity to examine which gene regions required ATX1 occupancy for TBP promoter binding. We observed that Flap or Selic treatments did not reduce TBP levels at the promoter regions of the ATX1-independent ACT7, TUBULIN6 (TUB6), and ACT12 genes (Figure 7B). More importantly, Flap or Selic treatments did not reduce TBP levels at the promoter regions of the ATX1-dependent WRKY70, NCED3, and LTP genes either (Figure 7B). This result indicates TBP recruitment did not require the presence of ATX1 within the transcribed regions of ATX1-dependent genes. For comparison purposes, the amounts of TBP and ATX1 at the promoters of these genes in wild-type and atx1 genotypes were also measured. TBP and ATX1 levels were decreased at the promoters of WRKY70, NCED3, and LTP in the atx1 genotype, while TBP levels were not diminished at the promoters of ACT7, TUB6, and ACT12 (Figure 7B). Together, these results demonstrate that TBP promoter recruitment at ATX1-regulated genes requires the presence of ATX1 at the promoter region, but not within the transcribed region.
DISCUSSION
This study provides new insights into the roles of ATX1 in the transcriptional regulation of three Arabidopsis genes that are directly targeted by ATX1. Collectively, our results showed that despite some similarities, ATX1 affects transcription by mechanisms different from those reported for yeast SET1 or mammalian MLL1. ATX1 plays dual roles, a plurality that has not been reported for yeast SET1 or mammalian MLL1. Specifically, ATX1 bound to the promoters and +300-bp regions of the three ATX1-regulated genes examined and exhibited two distinct roles in facilitating TBP and Pol II occupancy at promoters and in H3K4 trimethylation within the transcribed region. These roles were not clearly separable in an atx1 mutant, but could be uncoupled through chemical inhibition of CTD phosphorylation, which diminished ATX1 recruitment to the +300-bp regions without diminishing ATX1 recruitment to the promoter regions. Under these conditions, the peak of ATX1 binding in the genes occurred at the promoter regions, demonstrating that the peak of ATX1 occupancy at promoters was a valid observation and occurred independently of ATX1 accumulation at the +300-bp regions. The two different CDK7/CDK9 kinase inhibitors used have different chemical structures (Shapiro, 2006) but produced similar effects, which increases the validity of the results.
The NCED3 gene best demonstrated the two different roles of ATX1. The Flap inhibitor considerably reduced Ser5P levels on the CTD of Pol II bound to the NCED3 gene (Figure 6C). This caused ATX1 and H3K4me3 levels to decrease at the +300-bp region, consistent with a Ser5P requirement for ATX1 recruitment to the +300-bp region. However, TBP and ATX1 levels were not altered at the NCED3 promoter (Figure 7B). Most importantly, nearly normal levels of NCED3 transcripts were produced in the Flap-treated samples in contrast with the ~80% reduction in NCED3 transcripts in the atx1 mutant (Figure 4C). Thus, NCED3 expression was strongly dependent on ATX1 occupancy at its promoter but not on ATX1’s presence at the +300-bp region. The absence of ATX1 at the +300-bp region was associated with a loss of H3K4me3 in this region. These data illustrate that the functional role of ATX1 in regulating transcription was separable from its role in H3K4 trimethylation.
ATX1’s role at promoters was also supported by its occurrence in a protein complex with TBP. TBP, a strictly promoter-localized component of the PIC, and ATX1 were found within an immunoprecipitated protein complex (Figure 5A). The possibility that the coimmunoprecipitation of these proteins might be via a DNA fragment spanning separate locations is unlikely as the samples were not chemically cross-linked. Additionally, we performed this coimmunoprecipitation experiment in the presence or absence of DNase I and found that ATX1 and TBP were still coimmunoprecipitated when DNA levels were reduced by 200- to 1000-fold (see Supplemental Figure 3 online). We conclude that coimmunoprecipitation of ATX1 with TBP was due to their occurrence within a shared protein complex and was not via a DNA linkage mechanism.
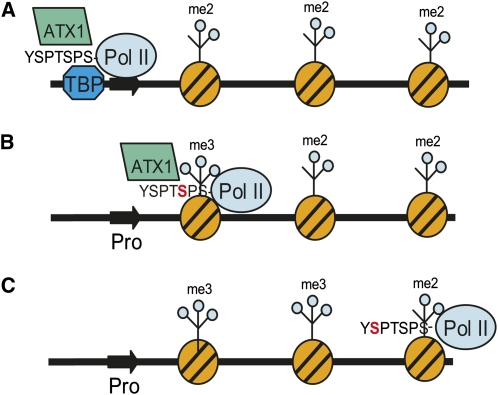
Based on the above data, we propose a model for how ATX1 functions at the promoter and +300-bp regions of the three ATX1-regulated genes (Figure 8). In the model, ATX1 first participates in a protein complex containing TBP to help initiate transcription (Figure 8A). CTD Ser5 phosphorylation occurs upon Pol II’s transition to transcription initiation/elongation states (Buratowski, 2009). ATX1 is then recruited to Ser5P-containing regions (with a peak around the +300-bp region of transcribed genes) to mediate methylation of H3K4me3 in a Ser5P-dependent manner (Figure 8B). Further transcriptional elongation results in a change in CTD phosphorylation status to predominantly the Ser2P form, releasing ATX1 due to a lack of binding to this form of the CTD (Figure 8C).
Figure 8.
Model of ATX1 Interactions at Two Locations in Target Genes.
A representation of an ATX1-dependent gene with its promoter (thick dark arrow) in a nucleosome-free region, and with the remainder of the gene complexed with nucleosomes (striped orange circles) containing H3K4me2 marks (me2), which are assumed to be the substrate for the trimethylating activity of ATX1 in this model, is shown.
(A) TBP, ATX1, and Pol II participate in the formation of a protein complex at the promoter, wherein the interaction of ATX1 with the nonphosphorylated form of the CTD of Pol II and additional undefined ATX1 interactions help stabilize this complex.
(B) Transcription elongation has moved Pol II to the +300-bp region of the transcribed gene, and Ser5 has become phosphorylated (red S in CTD consensus repeat YSPTSPS), recruiting ATX1 and facilitating trimethylation of H3K4 (me3) in this region.
(C) Continued transcription elongation by Pol II changes the phosphorylation status of the CTD to Ser2P (red S in CTD consensus repeat YSPTSPS). ATX1 has been released from Pol II as it does not bind to the Ser2P form of CTD.
In contrast with the ATX1-dependent transcription initiation suggested by our data, transcription initiation in yeast does not require SET1, as SET1/COMPASS is recruited subsequently to Pol II transcription initiation (Ng et al., 2003). The interaction between SET1 and the CTD is indirect, mediated by the Pol II–associated factor complex (Ng et al., 2003), whereas recruitment of ATX1 to the Ser5P form of the CTD could be directly mediated through the SET domain of ATX1. In the case of MLL1, loss of MLL1 affects Pol II occupancy at target genes, possibly through a mechanism involving binding of H3K4me3 by the PHD domain of the TAF3 subunit of TFIID (Vermeulen et al., 2007). Interestingly, Arabidopsis lacks a TAF3 subunit (Lawit et al., 2007) and yeast TAF3 lacks a PHD finger (Gangloff et al., 2001), indicating that TAF3 tethering of PIC to H3K4me3 nucleosomes is not a general mechanism in all organisms. In further support of this difference, treatment with Flap or Selic CDK7/CDK9 inhibitors reduced H3K4me3 levels without reducing TBP occupancy at the three ATX1-dependent promoters examined (Figure 7B). Importantly, this result also indicates that high H3K4me3 levels are not required for promoter accessibility. We conclude that ATX1, rather than H3K4 trimethylated nucleosomes, is critical for TBP and Pol II recruitment and/or stability at the promoters of ATX1-regulated genes in Arabidopsis. This direct coupling to the basal transcriptional machinery independently from TAF3-H3K4me3 anchoring is a previously unknown role for a histone H3K4-methyltransferase.
The presence of SET1/TRX-type proteins, H3K4me3, and Ser5P in 5′ regions of transcriptionally active genes has been observed in yeast, mammals, and plants (Ng et al., 2003; Milne et al., 2005; Kim et al., 2008). Although an earlier study has suggested that Ser5P was essential for transcription in yeast (Valay et al., 1995), recent evidence shows that phosphorylation of Ser5P is not required for transcription in yeast (Kanin et al., 2007). Our results in Arabidopsis are consistent with these more recent results in yeast, as inhibition of Ser5P/Ser2P did not interfere with TBP or ATX1 binding to the promoter regions.
H3K4me3 marks are recognized by chromatin remodeling factors facilitating transcription by altering the structure, composition, and positioning of nucleosomes, by components of the spliceosome, and by proteins involved in mRNA capping and stability (Ansari and Mandal, 2010). Recognition and binding to H3K4me3 has been traced to PHD domains or chromodomains present in these proteins (Ruthenburg et al., 2007). The PHD domain of the Arabidopsis ORC1 protein, which is not related to the trithorax protein family, has been demonstrated to bind H3K4me3 and affect transcription at target genes (de la Paz Sanchez and Gutierrez, 2009). The ePHD domain in ATX1 is related to the H3K4me3 binding PHD domain but has not been demonstrated to bind to H3K4me3. Our experiments unambiguously demonstrate that the degree of H3K4me3 modifications and mRNA production could be uncoupled in the case of NCED3 and ACT7. The fact that the WRKY70 and LTP transcript levels were reduced despite normal TBP and ATX1 occupancy at their promoters suggests that the reduced levels of one or more of the Ser2P, Ser5P, and H3K4me3 modifications did affect some aspect of downstream transcription elongation or mRNA processing for these genes. This suggests that different genes require different amounts of Ser2P, Ser5P, or H3K4me3 modifications for efficient transcript production. This is consistent with a role of these modifications in orchestrating posttranscriptional RNA processing (Phatnani and Greenleaf, 2006), in conjunction with a hypothesis that the threshold requirements for efficient expression vary for different genes.
The mechanisms by which ATX1 is targeted to specific DNA sequences are still unclear, as ATX1 does not have any known DNA binding motifs. The presence of ATX1 at two locations in ATX1-regulated genes might be facilitated by a single mechanism for recruiting ATX1 to target genes. Our hypothesis is that ATX1 is recruited to promoters in a protein complex containing TBP and that upon phosphorylation of Ser5 of the CTD of Pol II, ATX1 is recruited to the adjacent regions enriched in the Ser5P form of the CTD of Pol II (Figure 8). The key results supporting this hypothesis are that ATX1 binds to Ser5P and that recruitment of ATX1 to the promoter region can be separated from its recruitment to the +300-bp region when Ser5P formation was inhibited. We suggest that during promoter activation in Arabidopsis, the ability of ATX1 to participate in a protein complex with TBP and to bind to the nonphosphorylated, preinitiation state of the CTD of Pol II increases the occupancy of these proteins at promoters. However, these general interactions do not adequately explain the gene specificity of ATX1. One possible mechanism for specificity could rely on the considerable diversity that exists within the subunit composition of basal transcription factor complexes to facilitate their recognition of different core promoters (Müller et al., 2007; Juven-Gershon and Kadonaga, 2010). In this hypothesis, ATX1 binds to specific basal transcription factor complexes that interact with unique transcription factors to guide each type of complex to individual promoters. Additional research will be required to elucidate the mechanisms involved in ATX1’s newly revealed role in targeting specific promoter regions. The dual role of ATX1 in facilitating TBP and Pol II recruitment and in mediating H3K4 trimethylation distinguishes it from the roles reported for SET1 and MLL1.
METHODS
Genotypes, Plant Growth, and Treatments
The atx1-1 allele (Alvarez-Venegas et al., 2003), referred to here as atx1, contains a T-DNA insertion in its coding region between the DAST and ePHD domains (Alvarez-Venegas et al., 2003) and does not produce any detectable full-length ATX1 mRNA or protein (Alvarez-Venegas et al., 2006). Wassilewskija and atx1 plants were grown at 22°C with 12 h light for 20 d. Flavopiridol and Seliciclib solutions of 3 or 300 μM, respectively, were vacuum infiltrated into 3-week-old plants in soil. The infiltrated plants were grown for 6 h in the greenhouse to recover. For all NCED3 experiments, NCED3 transcription was activated by air drying for 1 h before tissue harvest.
Plasmid Constructs
An ATX1 cDNA (Alvarez-Venegas et al., 2006) was used to generate the gene fragments of ATX1 shown in Figure 2, and pGBKT7 and pGADT7 were from Clontech. Plasmids were constructed with the DNA primers and protocols described (see Supplemental Table 1 online). All cloned DNAs were confirmed by DNA sequencing.
Protein and Peptide Pull-Down Assays, Immunoprecipitations, and Immunoblots
Protein expression and purification were performed as described previously (Ding et al., 2007). Nuclear protein isolation from leaves and protein immunoprecipitations were performed as previously described (Serino and Deng, 2007). In brief, 3 to 5 g of leaves were ground in buffer (0.4 M sucrose, 10 mM Tris, pH 8.0, 5 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail [P9599, Sigma-Aldrich]) and filtered through Miracloth. After centrifugation, the pellet was suspended in buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1 mM PMSF, and protease inhibitor cocktail) and resuspended with a dounce homogenizer. After centrifugation, the supernatant was precleared with protein A (10002D; Invitrogen) or protein G magnetic beads (10003D; Invitrogen), and specific antibodies or control IgG serum were added for overnight incubation at 4°C. Antibody complexes were precipitated with protein A or protein G magnetic beads. The beads were washed with buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% Triton X-100, and protease inhibitor cocktail) and then boiled for 5 min in SDS loading buffer, and the proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Bio-Rad). Immunoblots were analyzed with antibodies to ATX1 (see Supplemental Figure 4 online); CTD (Abcam; ab817, lot 669648), the Ser2P form of Pol II CTD (Abcam; ab5095, lot 703307), the Ser5P form of Pol II CTD (Abcam; ab5131, lot 806890), trimethyl-H3K4 (Abcam; ab8580, lot 598382), H3 (Abcam; ab1791, lot 517990), the N terminus of Arabidopsis Pol II (Santa Cruz Biochemicals; sc-33754, lot E2406), or TBP (Abcam; ab52887, lot 347607).
For the CTD/SET pull-down assay with His-CTD as the soluble bait, GST beads were incubated with 2 μg of each GST fusion protein, washed, and then incubated with 3 μg of His-CTD protein overnight at 4°C. Mock controls used extracts prepared from either the His-Tag or GST vectors. The beads were washed five times (1× PBS buffer, pH 7.4, 1 mM PMSF, and 0.1% Triton X-100), and the remaining proteins eluted from the beads in SDS loading buffer, separated on a 10% SDS-PAGE gel, and analyzed by immunoblot with antibody to CTD (Abcam; ab817, lot 669648). For the CTD/SET domain pull-down assay with GST-SET as the soluble bait, the analogous procedure was followed, and GST-SET binding was detected with antibody to GST (Applied Biological Materials; G018, lot 5019).
The binding of the GST-SET domain proteins to synthetic CTD peptides was done similarly, except that 1.5 μg of biotinylated CTD peptides (Bio Basic) containing nonphosphorylated [(Y1S2P3T4S5P6S7)4] or the Ser2P [(Y1(p-S2)P3T4S5P6S7)4] or Ser5P [(Y1S2P3T4(p-S5) P6S7)4] phosphorylated forms of four repeats of the CTD heptamer consensus sequence were bound to 0.5 mg of streptavidin-coated beads (Dynabeads M280; Invitrogen) in 50 μL of high salt binding buffer (25 mM Tris-HCl, pH 8.0, 1 M NaCl, 1 mM DTT, 5% glycerol, and 0.03% Nonidet P-40) at 4°C for 2 h. The protein-bound beads were washed once with high salt binding buffer and twice with CTD binding buffer (25 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM DTT, 5% glycerol, and 0.03% Nonidet P-40) and finally resuspended in 50 μL of CTD binding buffer (Li et al., 2003). The beads were incubated with 3 μg of GST-SET protein or GST alone overnight at 4°C. The beads were washed five times (1× PBS buffer, pH 7.4, 1 mM PMSF, and 0.1% Triton X-100), and the remaining proteins eluted from the washed beads in SDS loading buffer. Samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Bio-Rad). GST-SET was detected by immunoblot analysis with antibody to GST (Applied Biological Materials; G018, lot 5019).
Reverse Transcription and Real-Time PCR
Total RNA isolation and reverse transcription with oligo(dT) (18418-012; Invitrogen) or random primers (48190-011; Invitrogen) were performed as described previously (Ding et al., 2007), and the amounts of individual genes were measured with gene-specific primers (see Supplemental Table 2 online). Real-time PCR analysis was performed with the cyclerIQ real-time PCR instrument (Bio-Rad) and SYBR Green mixture (Bio-Rad). The relative expression or amount of specific genes was quantitated with the 2−ΔΔCt calculation according to the manufacturer’s software (Bio-Rad; Livak and Schmittgen, 2001), where ΔΔCt is the difference in the threshold cycles and the reference housekeeping gene, which was ubiquitin for expression analyses or relative to input DNA for chromatin immunoprecipitation assays. rRNA was used as the internal control in expression analyses with the Flap or Selic inhibitors as RNA Polymerase I lacks a CTD. The mean threshold cycle values for the genes of interest were calculated from three replicates.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed according to manufacturer’s protocols (Clontech). Briefly, Saccharomyces cerevisiae strain Y190 was transformed with bait construct pGBKT-CTD and then transformed with pGADT7-ATX1, pGADT7-ATX1N, pGADT7-ATX1C, pGADT7-ATX1DH, or pGADT7-ATX1SET. Vectors without coding region inserts were used as negative controls. Yeast was scored for protein interactions by their ability to grow on SD medium lacking Trp, Leu, His, and adenine.
ChIP Assay
The ChIP assay was performed with a modified method (Ding et al., 2007). Briefly, 3 g of leaves were fixed with 1% formaldehyde for 10 min and quenched in 0.125 M glycine, and the leaves were ground in a mortar and pestle in buffer I (0.4 M sucrose, 10 mM Tris, pH 8.0, 5mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and filtered through Miracloth. After centrifugation, the pellet was extracted by buffer II (0.25 M sucrose, 10 mM Tris, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and then by buffer III (1.7 M sucrose, 10 mM Tris, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail). The nuclei were then lysed in lysis buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 1% SDS, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and the extract sonicated to fragment the DNA to a size range of 300 to 500 bp. After centrifugation, the supernatant was diluted by dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8.0, 167 mM NaCl, 0.1 mM PMSF, and protease inhibitor cocktail) and then precleared with protein A or protein G magnetic beads. Specific antibodies (described above) or control IgG serum were added to the precleared supernatants for an overnight incubation at 4°C. The antibody protein complexes were isolated by binding to protein A or protein G beads. The washed beads were heated at 65°C for 8 h with proteinase K to reverse the formaldehyde cross-linking and digest proteins. The sample was then extracted with phenol/chloroform and the DNA precipitated in ethanol and resuspended in water. Purified DNA was analyzed by real-time PCR with gene-specific primers (see Supplemental Table 2 online).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACT7, At5g09810; ACT12, At3G46520; LTP7, At2g15050; NCED3, At3g14440; TUB6, At5G12250; ubiquitin, At4g05320; and WRKY70, At3G56400.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Specificity of Ser2P and Ser5P Antibodies.
Supplemental Figure 2. Commercial Antibody to Mammalian TBP Recognizes Arabidopsis TBP.
Supplemental Figure 3. Coimmunoprecipitation of ATX1 and TBP Is Independent of DNA.
Supplemental Figure 4. Characterization of the Antibody to ATX1.
Supplemental Table 1. Plasmids Used.
Supplemental Table 2. PCR Primers for cDNA or ChIP.
Acknowledgments
We thank Heriberto Cerutti for critically reading the manuscript. The studies were supported by National Science Foundation Grant EPS-0701892 to Z.A. and M.F.
References
- Alvarez-Venegas R., Abdallat A.A., Guo M., Alfano J.R., Avramova Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2: 106–113 [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R., Avramova Z. (2001). Two Arabidopsis homologs of the animal trithorax genes: A new structural domain is a signature feature of the trithorax gene family. Gene 271: 215–221 [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R., Avramova Z. (2005). Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 33: 5199–5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R., Pien S., Sadder M., Witmer X., Grossniklaus U., Avramova Z. (2003). ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13: 627–637 [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R., Sadder M., Hlavacka A., Baluska F., Xia Y., Lu G., Firsov A., Sarath G., Moriyama H., Dubrovsky J.G., Avramova Z. (2006). The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. USA 103: 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari K.I., Mandal S.S. (2010). Mixed lineage leukemia: Roles in gene expression, hormone signaling and mRNA processing. FEBS J. 277: 1790–1804 [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Humphrey E.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L. (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99: 8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C.E., Minvielle-Sebastia L., Lee B.A., Keller W., Proudfoot N.J. (1998). Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301 [DOI] [PubMed] [Google Scholar]
- Buratowski S. (2009). Progression through the RNA polymerase II CTD cycle. Mol. Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C.I., Millar T., Finnegan E.J., Pogson B.J. (2009). Promoting gene expression in plants by permissive histone lysine methylation. Plant Signal. Behav. 4: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R.D., Heidemann M., Albert T.K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007). Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- de la Paz Sanchez M., Gutierrez C. (2009). Arabidopsis ORC1 is a PHD-containing H3K4me3 effector that regulates transcription. Proc. Natl. Acad. Sci. USA 106: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Lapko H., Ndamukong I., Xia Y., Al-Abdallat A., Lalithambika S., Sadder M., Saleh A., Fromm M., Riethoven J.-J., Lu G., Avramova Z. (2009). The Arabidopsis chromatin modifier ATX1, the myotubularin-like AtMTM, and the response to drought. Plant Signal. Behav. 4: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Wang X., Su L., Zhai J., Cao S., Zhang D., Liu C., Bi Y., Qian Q., Cheng Z., Chu C., Cao X. (2007). SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Murphy S. (2008). Cracking the RNA polymerase II CTD code. Trends Genet. 24: 280–288 [DOI] [PubMed] [Google Scholar]
- Fabrega C., Shen V., Shuman S., Lima C.D. (2003). Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol. Cell 11: 1549–1561 [DOI] [PubMed] [Google Scholar]
- Gangloff Y.G., Pointud J.C., Thuault S., Carré L., Romier C., Muratoglu S., Brand M., Tora L., Couderc J.L., Davidson I. (2001). The TFIID components human TAF(II)140 and Drosophila BIP2 (TAF(II)155) are novel metazoan homologues of yeast TAF(II)47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 21: 5109–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Olmedo F., Molina A., Alamillo J.M., Rodríguez-Palenzuéla P. (1998). Plant defense peptides. Biopolymers 47: 479–491 [DOI] [PubMed] [Google Scholar]
- Gomes N.P., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M. (2006). Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T., Kadonaga J.T. (2010). Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanin E.I., Kipp R.T., Kung C., Slattery M., Viale A., Hahn S., Shokat K.M., Ansari A.Z. (2007). Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl. Acad. Sci. USA 104: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., To T.K., Ishida J., Morosawa T., Kawashima M., Matsui A., Toyoda T., Kimura H., Shinozaki K., Seki M. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49: 1580–1588 [DOI] [PubMed] [Google Scholar]
- Kim M., Suh H., Cho E.J., Buratowski S. (2009). Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284: 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho E.J., Buratowski S. (2000). Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lawit S.J., O’Grady K., Gurley W.B., Czarnecka-Verner E. (2007). Yeast two-hybrid map of Arabidopsis TFIID. Plant Mol. Biol. 64: 73–87 [DOI] [PubMed] [Google Scholar]
- Lee Y.C., Park J.M., Min S., Han S.J., Kim Y.J. (1999). An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19: 2967–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Howe L., Anderson S., Yates J.R., III, Workman J.L. (2003). The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278: 8897–8903 [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2008). High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 20: 259–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. (1997). The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Milne T.A., Dou Y., Martin M.E., Brock H.W., Roeder R.G., Hess J.L. (2005). MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. USA 102: 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Demény M.A., Tora L. (2007). New problems in RNA polymerase II transcription initiation: Matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 282: 14685–14689 [DOI] [PubMed] [Google Scholar]
- Ng H.H., Robert F., Young R.A., Struhl K. (2003). Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Phatnani H.P., Greenleaf A.L. (2006). Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Pien S., Fleury D., Mylne J.S., Crevillen P., Inzé D., Avramova Z., Dean C., Grossniklaus U. (2008). ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Zeevaart J.A. (1999). The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 96: 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Hu C., Hinnebusch A.G. (2009). Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33: 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg A.J., Li H., Patel D.J., Allis C.D. (2007). Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008a). Dynamic and stable histone H3 methylation patterns at the Arabidopsis FLC and AP1 loci. Gene 423: 43–47 [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Yilmaz M., Le O., Hou G., Sadder M., Al-Abdallat A., Xia Y., Lu G., Ladunga I., Avramova Z. (2008b). The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Serino G., Deng X.W. (2007). Protein coimmunoprecipitation in Arabidopsis. Cold Spring Harb. Protoc. doi/10.1101/pdb.prot4683 [DOI] [PubMed] [Google Scholar]
- Shapiro G.I. (2006). Cyclin-dependent kinase pathways as targets for cancer treatment. J. Clin. Oncol. 24: 1770–1783 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. (2008). Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20: 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A., Matsubayashi S., Yamaguchi M., Uchimiya H., Umeda M. (2003). Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett. 534: 69–74 [DOI] [PubMed] [Google Scholar]
- Shukla A., Chaurasia P., Bhaumik S.R. (2009). Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell. Mol. Life Sci. 66: 1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M., Shimotohno A., Yamaguchi M. (2005). Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant Cell Physiol. 46: 1437–1442 [DOI] [PubMed] [Google Scholar]
- Valay J.G., Simon M., Dubois M.F., Bensaude O., Facca C., Faye G. (1995). The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol. 249: 535–544 [DOI] [PubMed] [Google Scholar]
- van Dijk K., Ding Y., Malkaram S., Riethoven J.J., Liu R., Yang J., Laczko P., Chen H., Xia Y., Ladunga I., Avramova Z., Fromm M. (2010). Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 10: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerappan C.S., Avramova Z., Moriyama E.N. (2008). Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi. BMC Evol. Biol. 8: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M., Mulder K.W., Denissov S., Pijnappel W.W., van Schaik F.M., Varier R.A., Baltissen M.P., Stunnenberg H.G., Mann M., Timmers H.T. (2007). Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Wang P., et al. (2009). Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D. (2005). WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Zhang X., Bernatavichute Y.V., Cokus S., Pellegrini M., Jacobsen S.E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]