Fig. 1.
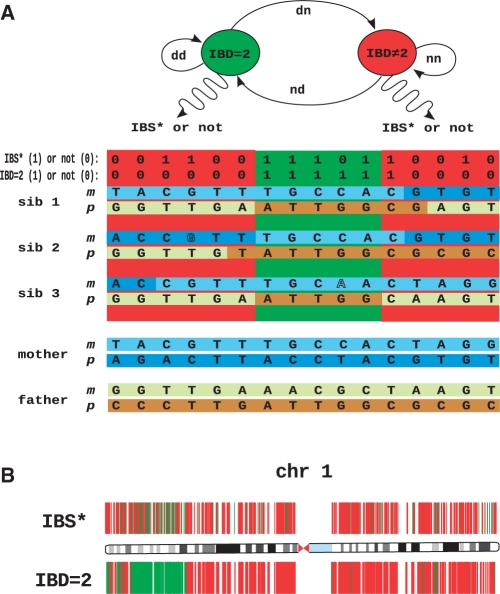
HMM to identify regions identical by descent in exome sequencing data. (A) In siblings affected with an autosomal recessive disorder, both the maternal and the paternal haplotypes surrounding the disease gene are identical by descent (IBD = 2). It is not possible to measure the IBD = 2 state directly, but only whether each sibling was called to the same homozygous or heterozygous genotype (referred to as IBS*). In this model, every genetic locus is either IBD = 2 or IBD ≠ 2 and the transition probabilities between these two states are defined by locus-specific transition rates, dd, nd, dn, nn. According to the HMM, these states emit genotypes that are IBS* or not, according to the appropriate probability distributions. Note that genotypes in IBD ≠ 2 may be IBS* by chance and genotypes in IBD = 2 may not be IBS* due to calling errors (displayed with outlined letters). The HMM and the observed exome sequence are used by the IBD = 2 classifying algorithm to identify regions of the genome that are IBD = 2. The disease gene must be located in such an IBD = 2 region. (B) Exome variant data of chromosome 1. Any chromosomal position that was called to a different genotype in at least one of three sibs on chromosome 1 with respect to the haploid reference sequence hg18 is depicted as a colored vertical line. In the upper panel, green indicates a genomic position that is IBS* in all three sibs, and red indicates ¬IBS* (non-IBS*). In the lower panel, green indicates genomic positions classified as IBD = 2, whereas red indicates IBD ≠ 2.