Abstract
In the title compound, [Mn(C6H4N5O)2(H2O)2]·2H2O, the MnII ion is situated on an inversion centre and is coordinated by the O and N atoms of two bis-chelating 5-(2-pyridyl-1-oxide)tetrazolate ligands and two O atoms of two water molecules in a distorted octahedral geometry. All the water H atoms are involved in O—H⋯N and O—H⋯O hydrogen bonds with uncoordinated water O atoms and tetrazole N atoms, which link the molecules into a three-dimensional network.
Related literature
For backgroud to tetrazolate derivatives in coordination chemistry, see: Jiang et al. (2007 ▶); Song et al. (2009 ▶); Zhang (2009) ▶. For related structures, see: Facchetti et al. (2004 ▶); Lin et al. (2005 ▶); Vrbova et al. (2000 ▶)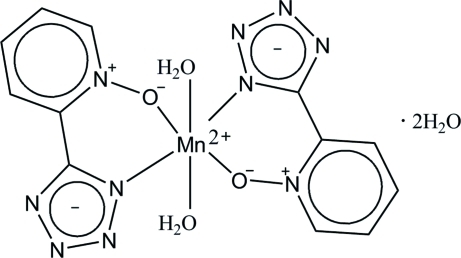
Experimental
Crystal data
[Mn(C6H4N5O)2(H2O)2]·2H2O
M r = 451.29
Monoclinic,

a = 6.4808 (13) Å
b = 12.034 (2) Å
c = 12.787 (4) Å
β = 116.24 (2)°
V = 894.5 (4) Å3
Z = 2
Mo Kα radiation
μ = 0.80 mm−1
T = 293 K
0.10 × 0.10 × 0.08 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2000 ▶) T min = 0.925, T max = 0.939
7432 measured reflections
1579 independent reflections
1102 reflections with I > 2σ(I)
R int = 0.115
Refinement
R[F 2 > 2σ(F 2)] = 0.078
wR(F 2) = 0.133
S = 1.14
1579 reflections
133 parameters
H-atom parameters constrained
Δρmax = 0.39 e Å−3
Δρmin = −0.38 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SAINT (Bruker, 2000 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811001620/lx2180sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811001620/lx2180Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3A⋯N2 | 0.88 | 2.15 | 3.010 (5) | 164 |
| O2—H2A⋯O3i | 0.84 | 2.01 | 2.756 (5) | 147 |
| O2—H2B⋯N3ii | 0.86 | 2.06 | 2.858 (5) | 154 |
| O3—H3B⋯N4ii | 0.82 | 2.10 | 2.917 (6) | 176 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
This work was supported by the Natural Science Foundation of Xuzhou Normal University (grant No. 09XLA06) and the National Natural Science Foundation of China (grant No. 21071121).
supplementary crystallographic information
Comment
Tetrazole as functional group plays an important role in coordination chemistry, medicinal chemistry and materials science applications (Song et al., 2009; Jiang et al., 2007; Zhang, 2009). It's interesting for the study of tetrazolate complexes to delineate the ways in which tetrazoles bind to metal centres. Here we report the structure of a novel substituted tetrazolato-metal complex, diaquabis(5-(2-pyridyl-1-oxide)tetrazolato)manganese(II) dihydrate.
The crystal structure of the title complex consists of the mononuclear manganese (II) unit [Mn(C6H4N5O)2(H2O)2], and two lattice water molecules (Fig. 1). In the mononuclear unit, manganese(II) ion is in a distorted octahedral environment, being six-coordinated by two N atoms and two O atoms from two bidentate 5-(2-pyridyl-1-oxide)tetrazolato-ligands, and two O atoms of two coordinated water molecules with Mn–O distances from 2.090 (4)Å to 2.209 (3) Å, Mn–N bond length of 2.255 (4)Å and O1–Mn1–N1 angle of 79.47 (14)°, which are comparable with the values observed in other metal-tetrazolate complexes (Vrbova et al., 2000; Lin et al., 2005; Facchetti et al., 2004). The pyridine and tetrazole rings are twisted against each other by 20.466 (190)°. In the crystal structure, all the water H atoms are involved in O–H···N and O–H···O hydrogen bonds with the solvate water O (O3W) and the tetrazole N (N2, N4) atoms. The interactions link the molecules into a three dimensional network (Table 1 and Fig. 2).
Experimental
A solution of 5-(2-pyridyl-1-oxide)tetrazole (32.6 mg, 0.2 mmol) and K2CO3 (13.8 mg, 0.1 mmol) in H2O (10 ml) was dropped slowly into a solution of Mn(ClO4).6H2O (36.2 mg, 0.1 mmol) dissolved in methanol (10 ml). The resulting brown suspension solution was stirred for 24 h at room temperature and filtered. Yellow crystals were separated from filtrate after about one month and collected for X-ray analysis (m.p. >573 K).
Refinement
H atoms were placed in calculated positions, with C–H = 0.93Å and O–H = 0.82-0.88 Å, and included in the final cycles of refinement using a riding model, with Uiso(H) = 1.2Ueq(parent atom).
Figures
Fig. 1.
The molecular structure of the title compound with the atomic numbering scheme. Displacement ellipsoids are drawn at the 30% probability level. [Symmetry code: (i) - x + 1, - y + 1, - z + 1. ]
Fig. 2.
A view of the O–H···O and O–H···N hydrogen bonds (dotted lines) in the crystal structure of the title compound.
Crystal data
| [Mn(C6H4N5O)2(H2O)2]·2H2O | F(000) = 462 |
| Mr = 451.29 | Dx = 1.676 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 7005 reflections |
| a = 6.4808 (13) Å | θ = 3.1–27.6° |
| b = 12.034 (2) Å | µ = 0.80 mm−1 |
| c = 12.787 (4) Å | T = 293 K |
| β = 116.24 (2)° | Block, yellow |
| V = 894.5 (4) Å3 | 0.10 × 0.10 × 0.08 mm |
| Z = 2 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 1579 independent reflections |
| Radiation source: fine-focus sealed tube | 1102 reflections with I > 2σ(I) |
| graphite | Rint = 0.115 |
| Detector resolution: 10.0 pixels mm-1 | θmax = 25.0°, θmin = 3.4° |
| φ and ω scans | h = −7→7 |
| Absorption correction: multi-scan (SADABS; Bruker, 2000) | k = −14→14 |
| Tmin = 0.925, Tmax = 0.939 | l = −15→15 |
| 7432 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.078 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.133 | H-atom parameters constrained |
| S = 1.14 | w = 1/[σ2(Fo2) + (0.0354P)2 + 1.2794P] where P = (Fo2 + 2Fc2)/3 |
| 1579 reflections | (Δ/σ)max < 0.001 |
| 133 parameters | Δρmax = 0.39 e Å−3 |
| 0 restraints | Δρmin = −0.38 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.5000 | 0.5000 | 0.5000 | 0.0282 (3) | |
| C1 | 0.4458 (9) | 0.2469 (4) | 0.4227 (4) | 0.0260 (12) | |
| C2 | 0.3159 (8) | 0.2280 (4) | 0.4903 (4) | 0.0276 (12) | |
| C3 | 0.2946 (10) | 0.1223 (5) | 0.5282 (5) | 0.0406 (15) | |
| H3 | 0.3648 | 0.0631 | 0.5099 | 0.049* | |
| C4 | 0.1752 (11) | 0.1017 (6) | 0.5912 (5) | 0.0531 (18) | |
| H4 | 0.1668 | 0.0303 | 0.6168 | 0.064* | |
| C5 | 0.0688 (10) | 0.1883 (6) | 0.6155 (5) | 0.0508 (17) | |
| H2 | −0.0165 | 0.1759 | 0.6568 | 0.061* | |
| C6 | 0.0860 (9) | 0.2922 (5) | 0.5800 (5) | 0.0407 (15) | |
| H1 | 0.0140 | 0.3510 | 0.5979 | 0.049* | |
| N1 | 0.5349 (7) | 0.3443 (3) | 0.4111 (3) | 0.0289 (10) | |
| N2 | 0.6423 (7) | 0.3211 (4) | 0.3443 (4) | 0.0338 (11) | |
| N3 | 0.6179 (8) | 0.2138 (4) | 0.3188 (4) | 0.0368 (12) | |
| N4 | 0.4943 (7) | 0.1654 (4) | 0.3670 (4) | 0.0346 (11) | |
| N5 | 0.2085 (7) | 0.3114 (4) | 0.5179 (4) | 0.0344 (11) | |
| O1 | 0.2069 (6) | 0.4131 (3) | 0.4819 (4) | 0.0491 (11) | |
| O2 | 0.2698 (6) | 0.5780 (3) | 0.3322 (3) | 0.0396 (10) | |
| H2A | 0.1657 | 0.5331 | 0.2904 | 0.048* | |
| H2B | 0.2775 | 0.6340 | 0.2925 | 0.048* | |
| O3 | 0.8363 (6) | 0.4852 (3) | 0.2336 (3) | 0.0447 (10) | |
| H3A | 0.7951 | 0.4450 | 0.2787 | 0.054* | |
| H3B | 0.7388 | 0.5334 | 0.2038 | 0.054* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0324 (7) | 0.0249 (6) | 0.0313 (7) | −0.0007 (6) | 0.0178 (5) | −0.0026 (6) |
| C1 | 0.028 (3) | 0.024 (3) | 0.025 (3) | 0.001 (2) | 0.011 (3) | 0.002 (2) |
| C2 | 0.027 (3) | 0.027 (3) | 0.027 (3) | 0.003 (2) | 0.011 (3) | −0.005 (2) |
| C3 | 0.053 (4) | 0.037 (4) | 0.040 (4) | −0.001 (3) | 0.028 (3) | −0.002 (3) |
| C4 | 0.069 (5) | 0.050 (4) | 0.046 (4) | −0.012 (4) | 0.030 (4) | 0.000 (3) |
| C5 | 0.046 (4) | 0.069 (5) | 0.038 (4) | −0.010 (4) | 0.019 (3) | 0.005 (4) |
| C6 | 0.033 (3) | 0.061 (4) | 0.035 (4) | 0.005 (3) | 0.021 (3) | −0.014 (3) |
| N1 | 0.030 (2) | 0.033 (3) | 0.026 (2) | 0.000 (2) | 0.016 (2) | −0.003 (2) |
| N2 | 0.032 (3) | 0.041 (3) | 0.032 (3) | −0.001 (2) | 0.018 (2) | −0.003 (2) |
| N3 | 0.038 (3) | 0.042 (3) | 0.034 (3) | 0.003 (2) | 0.019 (2) | −0.007 (2) |
| N4 | 0.043 (3) | 0.032 (3) | 0.035 (3) | −0.002 (2) | 0.024 (3) | −0.009 (2) |
| N5 | 0.035 (3) | 0.029 (3) | 0.034 (3) | 0.004 (2) | 0.011 (2) | 0.002 (2) |
| O1 | 0.043 (3) | 0.038 (2) | 0.076 (3) | −0.0019 (19) | 0.035 (2) | 0.000 (2) |
| O2 | 0.049 (2) | 0.038 (2) | 0.030 (2) | −0.0062 (19) | 0.016 (2) | 0.0060 (18) |
| O3 | 0.046 (2) | 0.040 (2) | 0.055 (3) | 0.005 (2) | 0.028 (2) | 0.012 (2) |
Geometric parameters (Å, °)
| Mn1—O1 | 2.090 (4) | C4—H4 | 0.9300 |
| Mn1—O1i | 2.090 (4) | C5—C6 | 1.351 (8) |
| Mn1—O2 | 2.209 (3) | C5—H2 | 0.9300 |
| Mn1—O2i | 2.209 (3) | C6—N5 | 1.369 (6) |
| Mn1—N1 | 2.255 (4) | C6—H1 | 0.9300 |
| Mn1—N1i | 2.255 (4) | N1—N2 | 1.348 (5) |
| C1—N4 | 1.329 (6) | N2—N3 | 1.324 (6) |
| C1—N1 | 1.344 (6) | N3—N4 | 1.341 (6) |
| C1—C2 | 1.467 (7) | N5—O1 | 1.306 (5) |
| C2—N5 | 1.353 (6) | O2—H2A | 0.8446 |
| C2—C3 | 1.390 (7) | O2—H2B | 0.8583 |
| C3—C4 | 1.363 (7) | O3—H3A | 0.8803 |
| C3—H3 | 0.9300 | O3—H3B | 0.8172 |
| C4—C5 | 1.359 (8) | ||
| O1—Mn1—O1i | 180.0 | C5—C4—C3 | 118.3 (6) |
| O1—Mn1—O2 | 85.11 (15) | C5—C4—H4 | 120.9 |
| O1i—Mn1—O2 | 94.89 (14) | C3—C4—H4 | 120.9 |
| O1—Mn1—O2i | 94.89 (15) | C6—C5—C4 | 120.4 (6) |
| O1i—Mn1—O2i | 85.11 (14) | C6—C5—H2 | 119.8 |
| O2—Mn1—O2i | 180.000 (1) | C4—C5—H2 | 119.8 |
| O1—Mn1—N1 | 79.47 (14) | C5—C6—N5 | 120.4 (5) |
| O1i—Mn1—N1 | 100.53 (14) | C5—C6—H1 | 119.8 |
| O2—Mn1—N1 | 92.20 (14) | N5—C6—H1 | 119.8 |
| O2i—Mn1—N1 | 87.80 (14) | C1—N1—N2 | 104.9 (4) |
| O1—Mn1—N1i | 100.53 (14) | C1—N1—Mn1 | 121.7 (3) |
| O1i—Mn1—N1i | 79.47 (14) | N2—N1—Mn1 | 133.4 (3) |
| O2—Mn1—N1i | 87.80 (14) | N3—N2—N1 | 108.6 (4) |
| O2i—Mn1—N1i | 92.20 (14) | N2—N3—N4 | 109.9 (4) |
| N1—Mn1—N1i | 180.000 (1) | C1—N4—N3 | 104.9 (4) |
| N4—C1—N1 | 111.7 (4) | O1—N5—C2 | 121.8 (4) |
| N4—C1—C2 | 122.4 (4) | O1—N5—C6 | 116.5 (5) |
| N1—C1—C2 | 125.9 (4) | C2—N5—C6 | 121.6 (5) |
| N5—C2—C3 | 116.4 (5) | N5—O1—Mn1 | 124.4 (3) |
| N5—C2—C1 | 122.3 (5) | Mn1—O2—H2A | 110.5 |
| C3—C2—C1 | 121.2 (5) | Mn1—O2—H2B | 135.8 |
| C4—C3—C2 | 122.8 (6) | H2A—O2—H2B | 111.6 |
| C4—C3—H3 | 118.6 | H3A—O3—H3B | 107.3 |
| C2—C3—H3 | 118.6 | ||
| N4—C1—C2—N5 | −160.1 (5) | O2i—Mn1—N1—N2 | −109.1 (4) |
| N1—C1—C2—N5 | 21.7 (8) | C1—N1—N2—N3 | −0.5 (5) |
| N4—C1—C2—C3 | 19.1 (8) | Mn1—N1—N2—N3 | 177.0 (3) |
| N1—C1—C2—C3 | −159.1 (5) | N1—N2—N3—N4 | 0.4 (5) |
| N5—C2—C3—C4 | −0.7 (8) | N1—C1—N4—N3 | −0.1 (6) |
| C1—C2—C3—C4 | 180.0 (5) | C2—C1—N4—N3 | −178.5 (4) |
| C2—C3—C4—C5 | 1.4 (9) | N2—N3—N4—C1 | −0.2 (5) |
| C3—C4—C5—C6 | −1.4 (9) | C3—C2—N5—O1 | −176.5 (5) |
| C4—C5—C6—N5 | 0.8 (9) | C1—C2—N5—O1 | 2.8 (7) |
| N4—C1—N1—N2 | 0.3 (6) | C3—C2—N5—C6 | 0.1 (7) |
| C2—C1—N1—N2 | 178.7 (5) | C1—C2—N5—C6 | 179.3 (5) |
| N4—C1—N1—Mn1 | −177.5 (3) | C5—C6—N5—O1 | 176.6 (5) |
| C2—C1—N1—Mn1 | 0.9 (7) | C5—C6—N5—C2 | −0.1 (8) |
| O1—Mn1—N1—C1 | −27.4 (4) | C2—N5—O1—Mn1 | −50.6 (6) |
| O1i—Mn1—N1—C1 | 152.6 (4) | C6—N5—O1—Mn1 | 132.7 (4) |
| O2—Mn1—N1—C1 | −112.1 (4) | O2—Mn1—O1—N5 | 146.5 (4) |
| O2i—Mn1—N1—C1 | 67.9 (4) | O2i—Mn1—O1—N5 | −33.5 (4) |
| O1—Mn1—N1—N2 | 155.5 (4) | N1—Mn1—O1—N5 | 53.3 (4) |
| O1i—Mn1—N1—N2 | −24.5 (4) | N1i—Mn1—O1—N5 | −126.7 (4) |
| O2—Mn1—N1—N2 | 70.9 (4) |
Symmetry codes: (i) −x+1, −y+1, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3A···N2 | 0.88 | 2.15 | 3.010 (5) | 164 |
| O2—H2A···O3ii | 0.84 | 2.01 | 2.756 (5) | 147 |
| O2—H2B···N3iii | 0.86 | 2.06 | 2.858 (5) | 154 |
| O3—H3B···N4iii | 0.82 | 2.10 | 2.917 (6) | 176 |
Symmetry codes: (ii) x−1, y, z; (iii) −x+1, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LX2180).
References
- Bruker (2000). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Facchetti, A., Abbotto, A., Beverina, L., Bradamante, S., Mariani, P., Stern, C. L., Marks, T. J., Vacca, A. & Pagani, G. A. (2004). Chem. Commun. pp. 1770–1771. [DOI] [PubMed]
- Jiang, T., Zhao, Y.-F. & Zhang, X.-M. (2007). Inorg. Chem. Commun. 10, 1194–1197.
- Lin, P., Clegg, W., Harrington, R. W. & Henderson, R. A. (2005). Dalton Trans. pp. 2388–2394. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Song, W.-C., Li, J.-R., Song, P.-C., Tao, Y., Yu, Q., Tong, X.-L. & Bu, X.-H. (2009). Inorg. Chem. 48, 3792–3799. [DOI] [PubMed]
- Vrbova, M., Baran, P., Boca, R., Fuess, H., Svoboda, I., Linert, W., Schubert, U. & Wiede, P. (2000). Polyhedron, 19, 2195–2201.
- Zhang, L. (2009). Acta Cryst. E65, m871–m872. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811001620/lx2180sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811001620/lx2180Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




