Figure 1.
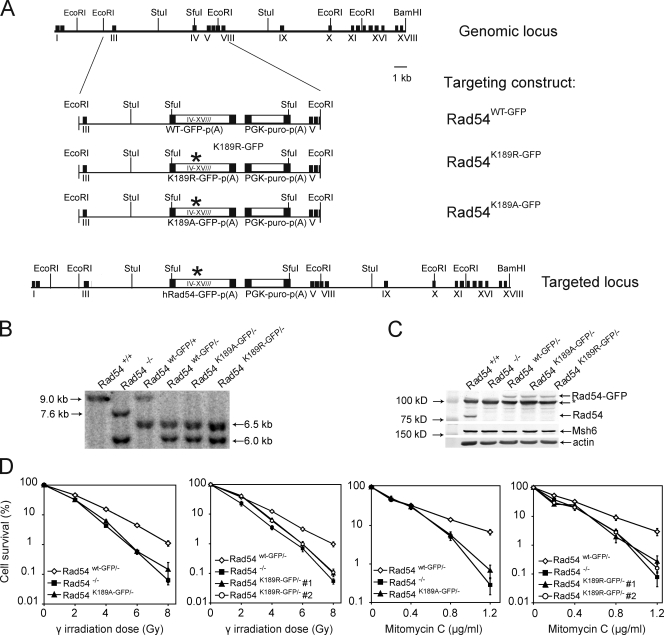
Characterization of mouse ES cells carrying ATPase-defective Rad54–GFP alleles. (A) Schematic representation of the mouse Rad54 locus and the gene-targeting constructs. The top line represents a 30-kb portion of endogenous Rad54 locus, where black boxes indicate exons I–XVIII. The middle line shows the linearized targeting construct, containing the human RAD54 cDNA sequence spanning exons IV–XVIII fused to the GFP coding sequence. The K189R and K189A mutations in the Walker A ATPase domain are indicated by the asterisks. The construct contains a gene encoding for puromycin resistance as a selectable marker. The targeting construct will replace the regions between exons III and VIII when correctly integrated to generate the targeted allele, as shown in the targeted locus. Homologous integration results in the expression of full-length, GFP-tagged Rad54 from its endogenous promoter. (B) DNA blot analysis of ES cells carrying the knockin contructs. DNA blot analysis was performed using genomic DNA purified from puromycin-resistant clones and digested with StuI. Detection of bands was performed using a probe that recognized exons VII/VIII. Restriction of the wild-type allele by StuI, (indicated by “+”), yields a 9.0-kb band after hybridization with an exon VII/VIII probe. Diagnostic bands for the neomycin-resistant knockout alleles, indicated by “−“, are 7.6 kb for a hygromycin-resistant allele and 6.0 kb for a neomycin-resistant allele. Knockin alleles are characterized by a doublet of bands ∼6.5 kb. (C) Immunoblot analysis of proteins produced by the Rad54–GFP knockin and -out alleles. Whole cell extracts of ES cells with the indicated genotypes were probed with affinity purified anti–human Rad54 antibodies. The position of Rad54 and Rad54–GFP are indicated. The arrowhead indicates a nonspecific signal. Probing against Msh6 and actin was used to confirm equal protein loading. (D) Ionizing radiation and mitomycin C survivals. ES cells of the indicated genotypes were tested for their ability to survive treatments with increasing doses of ionizing radiation (γ irradiation) or mitomycin C using clonogenic survival assays. The assays were performed in triplicate and the error bars indicated the standard error of the mean.