Abstract
Optimization of nonviral gene delivery typically focuses on the design of particulate carriers that are endowed with desirable membrane targeting, internalization, and endosomal escape properties. Topographical control of cell transfectability, however, remains a largely unexplored parameter. Emerging literature has highlighted the influence of cell-topography interactions on modulation of many cell phenotypes, including protein expression and cytoskeletal behaviors implicated in endocytosis. Using high-throughput screening of primary human dermal fibroblasts cultured on a combinatorial library of microscale topographies, we have demonstrated an improvement in nonviral transfection efficiency for cells cultured on dense micropit patterns compared to smooth substrates, as verified with flow cytometry. A 25% increase in GFP+ cells was observed independent of proliferation rate, accompanied by SEM and confocal microscopy characterization to help explain the phenomenon qualitatively. This finding encourages researchers to investigate substrate topography as a new design consideration for the optimization of nonviral transfection systems.
1. Introduction
The promise of gene medicine is intimately linked to the efficiency of nonviral transfection. Mechanistic understanding of the nonviral gene delivery process remains incomplete, and so is the structure-function relationship of nonviral vectors. Currently, particulate parameters of DNA nanocomplexes cannot fully account for the differences in nonviral transfection efficiency observed between various gene delivery systems (reviewed in [1]). At the same time, substrate parameters including stiffness [2, 3], co-presentation of adsorbed vectors with ECM and serum proteins [4–6], and surface chemistry [7, 8] have been demonstrated to strongly influence the uptake and expression of nonviral vectors, though comparatively little focus has been placed on their design. These differences are often attributed to vector-substrate interactions - using substrates as depots for the capture and/or controlled release of nonviral gene carriers, thereby increasing their local concentration at cell surfaces [9–11]. However, increasing importance is being placed on biochemical and physical cell-substrate interactions, which may prime cells to become more readily transfected [3, 12, 13]. For example, collagen-coated surfaces inhibit transfection of mesenchymal stem cells (MSCs) following bolus delivery of polyplexes compared to those cultured on uncoated surfaces, whereas fibronectin coating increases transgene expression by enhancing clathrin-mediated endocytosis [12]. Physically, stiffer substrates support more efficient transfection, attributed to enhanced proliferation on hard surfaces [3], which is believed to increase nuclear accessibility to nonviral vectors during cell division [14].
Substrate topography may also be playing a role in the transfection process, through cell-substrate and/or vector-substrate interactions. Patterned topography has been observed to engender a number of phenotypic changes in cells, many of which with implications in the delivery of nonviral vectors that have yet to be investigated. Cells interact with their substrates through integrins, a family of transmembrane receptors that bind to an assortment of extracellular matrix proteins. Integrin binding and clustering initiates the assembly of focal adhesion complexes [15]. Substrate topography and chemistry alter the amount and conformation of integrin ligands [16–20], and may form the general basis for many if not all subsequent topographical effects on cell behavior [19–21].
Altered cell spreading is one of the most often-observed effects of culture on nano- and microtopographic surfaces. The heterogeneous presentation of extracellular matrix proteins adsorbed to topography supports varying degrees of integrin engagement, leading to varying degrees of cell spreading [15–17] nuclear deformation [21–24], and subsequent changes in genomic expression profiles [24, 25]. The act of integrin engagement and spreading itself may have an effect on the clathrin- and caveolae-mediated uptake of nonviral vectors, two of the primary routes along which particulate gene carriers are delivered to the nucleus [26–29]. This rationale is supported by studies describing molecular links between cell substrates, focal adhesions, and the cytoskeletal and endocytic machineries [30–35].
Observing the influence of topographical cues on many cell phenotypes including proliferation [36–38], spreading, cytoskeletal organization, and endocytosis, we hypothesize that substrates with micropatterned topographies may also influence the transfectability of interacting cells. In this study, we investigated the transfection of normal human dermal fibroblasts (NHDFs) by GFP-encoding lipoplexes on a topographical library comprising 160 patterns of square and/or circular pit geometries, systematically varied with respect to size, spacing, and arrangement in the 1–6 μm range. Morphology and proliferation were also documented to further examine the observed differences in transfection efficiency. Patterns with elevated transfection levels on the library (3×3 mm2 each pattern) were replicated as large (2×2 cm2) single-pattern substrates for analysis by flow cytometry, luciferase assay, and characterization with SEM and confocal microscopy.
2. Materials and Methods
2.1 Preparation of 13 × 13 pattern topographical library
Topographical libraries (arrays) were prepared as described in [39], using a standard lithography process. Briefly, a resist without hard bake was etched with Cl2, HBr and NF3 to produce sidewall angles of 85° with etch rate non-uniformity of 2–3% (max–min). After removal of the resist and cleaning, the surface was sputter- coated with 100 nm of tantalum at a rate of 50 nm min−1, producing a 13 × 13 master library of post topographies. To create substrates for cell culture, Sylgard 184 polydimethylsiloxane (PDMS) (Dow Corning, Midland, MI) was mixed at a curing agent/base ratio of 1.05/10 w/w, degassed in a vacuum chamber, cast onto the metal master, and cured overnight at 47°C. PDMS arrays were peeled from the metal master and used for cell culture. These arrays were composed of 10 distinct pit morphologies (A–J) with: pit size (X) and edge-to-edge spacing (Y) iterated combinatorially through values of 1, 2, 4, and 6 μm (Figure 1), a uniform depth of 2.4 μm, and with 3×3 mm2 patterned area for each. Pattern K did not support robust NHDF adhesion, and was excluded from all analyses. Smooth control regions were present in the middle of the array, and in the boundaries between patterns. Large area (2×2 cm2) metal masters of the single patterns F(X,Y) = F(1,4) and F(4,1) were also produced following the same lithographical process to facilitate analysis by flow cytometry and luciferase assay. Pitted PDMS replicates of these “single-pattern” substrates, as well as smooth PDMS substrates produced from smooth silicon wafers, were stamped into 2 cm2 circular disks, affixed to the bottom of 24-well tissue culture plates (Falcon, Heidelberg, Germany) with 50 μL PDMS, and cured overnight.
Figure 1.
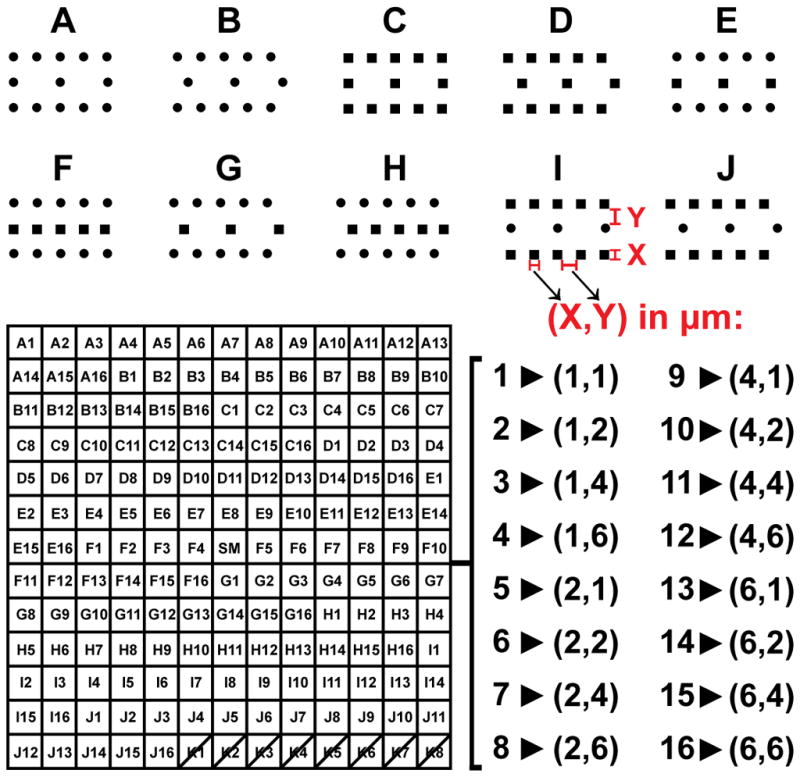
Topographical library details: 10 pit morphologies (A–J) were replicated with 16 different combinations of size (X, 1–6 μm) and spacing (Y, 1–6 μm), giving 160 unique patterned PDMS substrates for cell growth, each with a uniform pit depth of 2.4 μm. Smooth regions were present in the center of the array (SM), and in the regions between patterns. Pattern K was excluded from all analyses due to poor cell attachment.
2.2 Cell culture
Adult normal human dermal fibroblasts (NHDFs) (Lonza, Basel, Switzerland) were cultured in high glucose Dulbecco’s Modified Eagle’s Medium (GIBCO 11960-044) (Invitrogen, Carlsbad, CA) supplemented with 20% Premium Select FBS (Atlanta Biologicals, Lawrenceville, GA), 25 μg mL−1 gentamicin (Invitrogen), and 1× GlutaMAX, non-essential amino acids, sodium pyruvate, and β-mercaptoethanol (Invitrogen), at 37°C and 5% CO2. NHDFs were passaged a maximum of six times prior to experimentation. PDMS substrates were washed with 70% ethanol and air-dried twice, then washed with sterile deionized (DI) water and air-dried twice again. 25 μg mL−1 human plasma fibronectin (BD, Franklin Lakes, NJ) in DI water was adsorbed to PDMS substrates for 1 hour at room temperature (RT) before seeding NHDFs at a density of 7,500 cells cm−2 for quantitative analyses, and 3,750 cells cm−2 for visualization by SEM and confocal microscopy.
2.3 Plasmid DNA
pmaxGFP (Amaxa, Cologne, Germany) and VR1255 (Vical, San Diego, CA) plasmids expressing green fluorescent protein (GFP) and luciferase reporters under control of the CMV promoter were propagated in Escherichia coli DH5α and purified with the EndoFree Plasmid Giga Kit (Qiagen, Hilden, Germany). Plasmid DNA concentrations were quantified by measurement of absorbance at 260 nm with a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Waltham, MA).
2.4 Transfection protocol
24 hours after seeding on topographical libraries or single-pattern substrates, NHDFs were transfected with a 2:1 ratio of Lipofectamine 2000 (Invitrogen) volume (μL) to pmaxGFP or VR1255 DNA mass (μg) at a dose of 1 μg plasmid cm−2 in serum- and antibiotic-free OptiMEM (Invitrogen), according to the manufacturer’s protocol. Transfection medium was replaced with complete medium 4 hours after the onset of transfection. Mass ratio and DNA dose were selected based on a preliminary optimization of luciferase expression levels in NHDFs cultured on smooth substrates (results not shown).
2.5 Proliferation assay
Proliferation of cells on topographical libraries was quantified with the BrdU (5-bromo-2-deoxyuridine) incorporation assay, which labels the nuclei of cells that have undergone mitosis. Following removal of the transfection medium, cells were incubated for 20 hours with 15 μM BrdU (Invitrogen) in complete media. Cells were then washed briefly with Cellgro phosphate-buffered saline (PBS) containing Ca2+ and Mg2+ (Mediatech, Washington, DC), and fixed with 4% paraformaldehyde (PFA) (EMS, Hatfield, PA) at 4°C for 30 minutes. Genomic DNA was denatured with 2M HCl (Sigma-Aldrich, Saint Loius, MO) for 10 minutes at RT and 20 minutes at 37°C, immediately followed by a 5 minute wash with 0.1 M borate buffer (Thermo Scientific), and a 30 minute RT incubation in blocking buffer containing 0.2% Triton X-100, 3% w/v BSA, and 10% goat serum (Sigma-Aldrich). BrdU incorporation was detected by 12 hour incubation at 4°C with anti-BrdU-AF594 mAb (1:100 dilution, Invitrogen), and 15 minute RT DAPI counterstain (1:5000, Invitrogen). Between each staining step, cells were washed three times for five minutes at RT with PBS containing 0.2% Triton X-100 and 1% BSA. An example of the BrdU staining is provided in Supplementary Figure 1.
2.6 Automated fluorescence imaging and analysis of topographical library
24 hours after the onset of transfection, NHDFs were washed briefly with PBS containing Ca2+ and Mg2+, and fixed with 4% PFA (EMS) at 4°C for 30 minutes. Cell spreading was visualized with a 30 minute RT phalloidin-AF594 (1:200, Invitrogen) stain, and nuclei with 15 minute DAPI (1:5000, Invitrogen) stain at RT, with three five-minute PBS washes before and after each. Following staining, arrays were mounted face-down on Nunc single-well OmniTrays (Thermo Scientific) with Fluoro-Gel (EMS).
Transfection efficiency (GFP, green), cell spreading (phalloidin, red), and cell number (DAPI, blue) data were collected from three independent arrays, while proliferation data (BrdU, red; DAPI, blue) were collected from another three, using a Nikon Eclipse TE2000-U fluorescence inverted microscope fitted with appropriate filters and ProScanII robotic stage and autofocus controllers (Prior). Each array was scanned automatically using NIS-Elements software (Nikon, Tokyo, Japan), with autofocus performed in every field, and a constant exposure time for each channel within a given array, producing large high-resolution composite images for each fluorophore.
Adobe Photoshop CS3 (Adobe Systems, San Jose, CA) was used to overlay a black grid onto an image of an array, outlining the boundaries of each patterned region within that array (Supplementary Figure 2). This masked image was then used to define regions of interest (ROIs) in ImageJ (NIH). First, a threshold was set to differentiate the background in each region from the black mask. The “Analyze particles” command was then used to detect and store each patterned area as a unique ROI. The original (unmasked) images were then re-thresholded and analyzed with a custom ImageJ macro, which sequentially stepped through each ROI and utilized the “Analyze particles” command to count the number of BrdU+ nuclei, DAPI+ nuclei, and GFP+ cells, or quantified the area fraction of each region stained with phalloidin. Particles that were significantly smaller than cells or nuclei were excluded from the relevant quantifications with the “Particle size” setting within “Analyze particles”.
2.7 Flow cytometry of cells on single-patterned substrates
24 hours after the onset of transfection with pmaxGFP-containing complexes (48 hours after cell seeding), NHDFs cultured on F(1,4), F(4,1), and smooth PDMS substrates in quadruplicate were washed briefly with PBS containing Ca2+ and Mg2+, released from substrate surfaces with 0.25% Trypsin-EDTA (Invitrogen), centrifuged, and resuspended in propidium iodide (PI) (1:1000, Invitrogen) to stain dead cells. Cells were then filtered through 40 μm nylon cell strainers (BD) and analyzed with a BD FACSCanto II flow cytometer. Single-positive PI+ and GFP+ cells were used to adjust spectral compensation. Non-transfected NHDFs cultured on smooth PDMS served as negative control, with 1% of these cells considered GFP+.
2.8 Luciferase assay
24 hours after the onset of transfection with VR1255-containing complexes, NHDFs cultured on F(1,4), F(4,1), and smooth PDMS substrates in triplicate were washed briefly with PBS containing Ca2+ and Mg2+, and lysed with: 500 μL Glo Lysis Buffer (Promega, Fitchburd, WI), a −80°C freeze-thaw cycle, and scraping. Non-transfected cells served as negative control. 100 μL of each lysate was then combined with 100 μL Steady-Glo Luciferase Assay Substrate (Promega) in an opaque 96-well plate (Thermo Scientific). Luminescence signals were normalized to the total dsDNA content in each lysate measured by Quant-iT PicoGreen assay (Invitrogen), according to the manufacturer’s protocol. A standard curve of cell number to PicoGreen signal was linear in the required dynamic range. Luminescence integrated over 7 seconds (luciferase) and fluorescence (PicoGreen) values were collected at RT following 5 minute incubations using a BMG Labtech FLUOStar Optima plate reader.
2.9 Confocal laser scanning microscopy
To visualize NHDFs protruding into pits, the cells were stably transduced with fluorescent cytoskeletal fusion proteins Lifeact-mRFPRuby (F-actin) [40] and α-actinin-GFP [41] (Addgene plasmid 11908) which were previously cloned into the lentiviral vector FUGW, and delivered with replication-deficient 2nd generation lentiviruses (packaging vectors psPAX2 (Addgene plasmid 12260) and pMD2.G (Addgene plasmid 12259)). Lentiviral transduction was used in place of immunohistochemistry to minimize sample handling. The stably-transduced NHDFs were cultured on patterns F(1,4) and F(4,1) for 24 hours, washed with PBS containing Ca2+ and Mg2+, fixed for 20 minutes in 4% PFA at 4°C, mounted face-down in Fluoro-Gel (EMS) on #1.5 coverslips (VWR, West Chester, PA), and visualized with the 63× oil immersion objective of a Zeiss LSM 510 inverted confocal microscope.
2.10 SEM
Cells cultured on single-pattern substrates were washed briefly with PBS containing Ca2+ and Mg2+, and dehydrated with a graded ethanol series. The cell samples were then dried with a Pelco CPD2 critical point drier, and sputter coated with a 10 nm layer of gold using a Denton Desk IV Vacuum Sputter Unit. Single-pattern substrates without cells were cleaned with 70% ethanol, freeze-fractured with liquid nitrogen, and sputter-coated with 10 nm of gold. All samples were then examined with an FEI XL30 scanning electron microscope.
2.11 Statistics
Statistical treatment and graphing of data were performed using Prism (GraphPad Software, La Jolla, CA). For comparisons of multiple pairs, ANOVA with Tukey post-test was performed, with p-values < 0.05 considered significant. For post-normalization comparisons to smooth substrates, one-sample t-test was performed, with p-values < 0.05 considered significant. Error bars represent the standard deviation of three independent experiments.
3. Results
3.1 Transfection efficiency, morphology, and proliferation of fibroblasts on topographical libraries
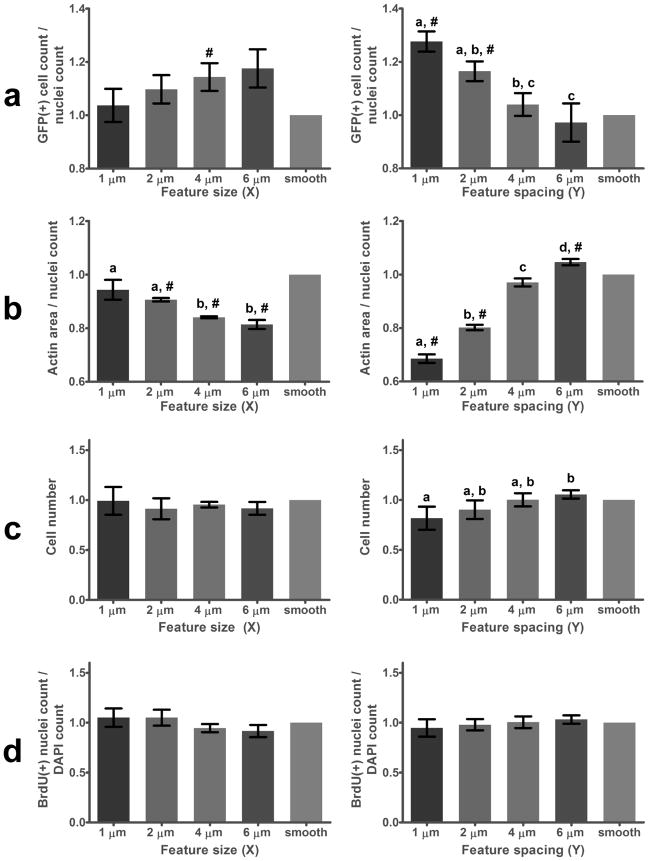
High-throughput parallel screening of 160 pitted topographies revealed a significant 25% enhancement of GFP transfection efficiency for cells cultured on patterns with small interfeature spacings (Y = 1, 2 μm) compared to smooth regions (Figure 2a). This enhancement decreased and disappeared as the pits became spaced further apart; that is, as they more closely approximated a smooth surface. Regions with large pits (X) increased transfection efficiency more modestly, while patterns with small pits supported identical transfection efficiencies as smooth regions. Pit morphologies A–J had no significant impact on transfection efficiency (results not shown). Topography-mediated enhancement of transfection was significant compared to smooth regions within each array, as well as between the averages of three normalized arrays presented in Figure 2. Examples of these differences are visible in representative widefield fluorescence images presented in Figure 3.
Figure 2.
(a) Transfection efficiency, (b) spreading, (c) cell number, and (d) proliferation of NHDFs cultured on topographical PDMS libraries normalized to values taken from smooth regions in each array. Data are grouped across pit morphologies A–J by pit diameter (X, left), and pit spacing (Y, right). Letters denote significance between columns and #’s denote significance compared to smooth regions. (a) Small interfeature spacing (Y = 1μm) produced a 25% increase in transfection efficiency compared to smooth regions, while feature size (X) had a less-significant impact. (b) Pits spaced far apart (large Y), and those with small diameters (small X), supported the highest levels of spreading. (c) Feature size (X) had no detectable effect on cell number whereas patterns with large interfeature spacing (Y = 6 μm) contained 25% more cells than those with small interfeature spacing (Y = 1 μm). (d) No significant dependence of proliferation on feature spacing was detected, suggesting increased cell number on features with large interfeature spacing may reflect increased initial attachment.
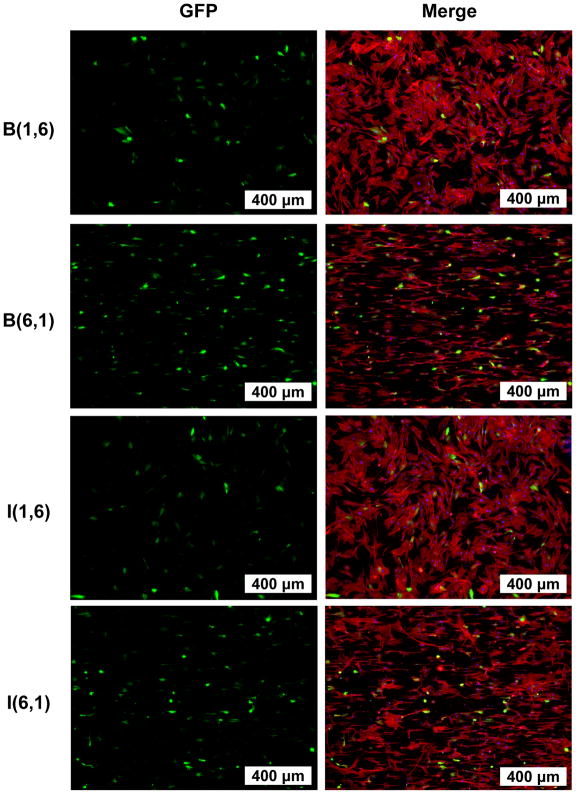
Figure 3.
Representative fluorescence microscopy images of NHDFs transfected on topographical PDMS libraries, with visible GFP transgene (green), actin (phalloidin stain, red), and nuclei (DAPI, blue). Patterns with small interfeature spacing (B(6,1) and I(6,1)) supported higher transfection efficiency, greater alignment, and less spreading than those with large interfeature spacing (B(1,6) and I(1,6)).
Cell spreading was quantified as the area occupied by actin on each pattern normalized by the number of cells in that region (Figure 2b). Spreading showed a trend opposite that observed for transfection efficiency; NHDFs on large, closely spaced pits were significantly less spread than those on small, distal pits, and on smooth regions. Regions with large, closely-spaced pits were also observed to encourage cell alignment. The strong influence of pitted topography on NHDF morphology is also evident in Figure 3.
Pit size had no effect on the number of cells in each patterned region after transfection. However, features with closely-spaced pits had slightly but significantly fewer cells than those with large interfeature distances (Figure 2c). No combination of pit size or spacing supported a significantly different number of cells than found on smooth regions. No significant differences in proliferation rates were observed for NHDFs cultured on topographical libraries (Figure 2d).
3.2 Flow cytometry on single-patterned substrates
Without normalization, an average of 55.0 ± 3.5% NHDFs cultured on smooth PDMS were GFP-positive following nonviral transfection, whereas an average of 68.3 ± 2.7% of cells cultured on pattern F(4,1) and 57.8 ± 3.6% on pattern F(1,4) were successfully transfected across all three experiments (95% confidence intervals). Normalization to the transfection efficiency of cells on smooth PDMS in each experiment gives a significant 24.4% average enhancement of transfection efficiency for cells cultured on F(4,1), compared to F(1,4) and smooth substrates (Figure 4). Representative flow cytometry data are provided in Supplementary Figure 3.
Figure 4.
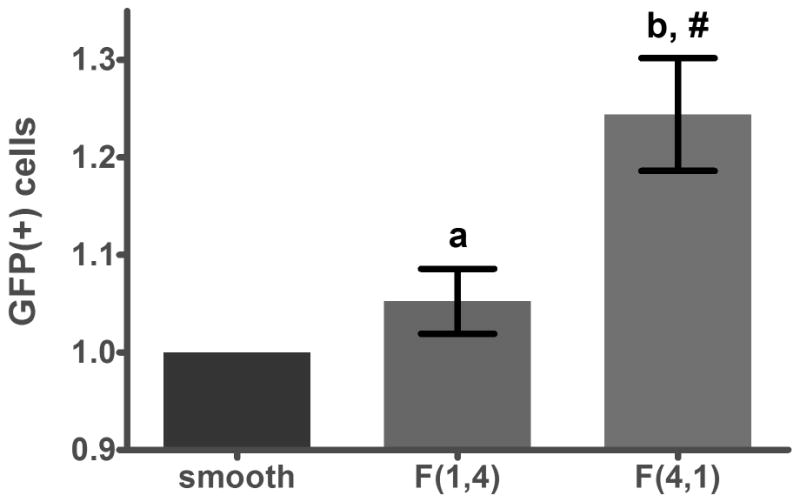
Flow cytometry corroborates the results presented in Figure 3a; F(4,1), a pattern with small interfeature spacing (1 μm), supported 25% higher transfection efficiency than smooth PDMS and F(1,4), a substrate with large interfeature spacing (4 μm).
3.3 Luciferase assay
Luciferase activity normalized to total cellular DNA was not significantly different for NHDFs transfected on smooth, F(1,4), or F(4,1) patterned topographies (data not shown). Cells transfected with luciferase were 4,000 times more luminescent than no-transfection negative controls.
3.4 SEM
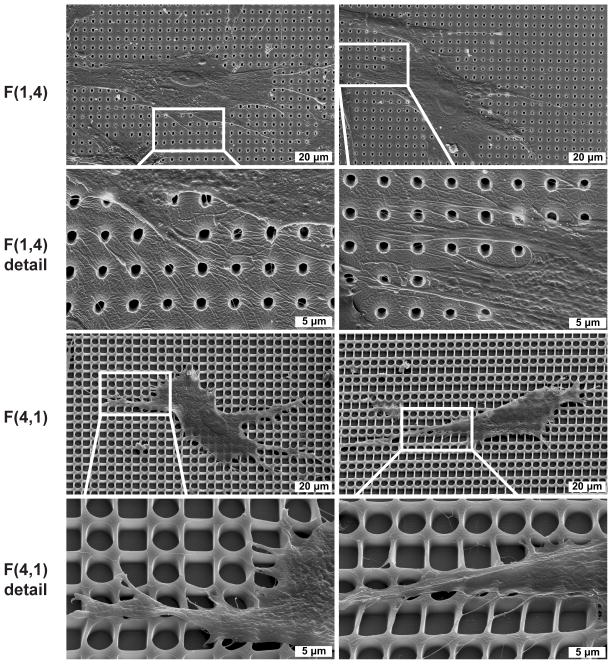
Inspection of NHDFs cultured on pitted topography with SEM revealed NHDFs cultured on pattern F(4,1) were less spread, and more aligned than those on pattern F(1,4) (Figure 5). The cells were also observed to drastically deform the walls of the 4 μm pits, sometimes with a single thin membrane process. For both patterns, cells could be seen extending portions of their membrane over the edge of open pits. Freeze fracture of the patterns revealed a uniform pit depth of 2.4 μm (Supplementary Figure 4).
Figure 5.
SEM of cells interacting with pitted topography, imaged with 30° tilt. NHDFs cultured on F(1,4) are more spread than those cultured on F(4,1). Regions of the cell membrane were stretched over open pits on both patterns. Cells cultured on F(4,1) were observed to dramatically deform pit sidewalls.
3.5 Confocal microscopy
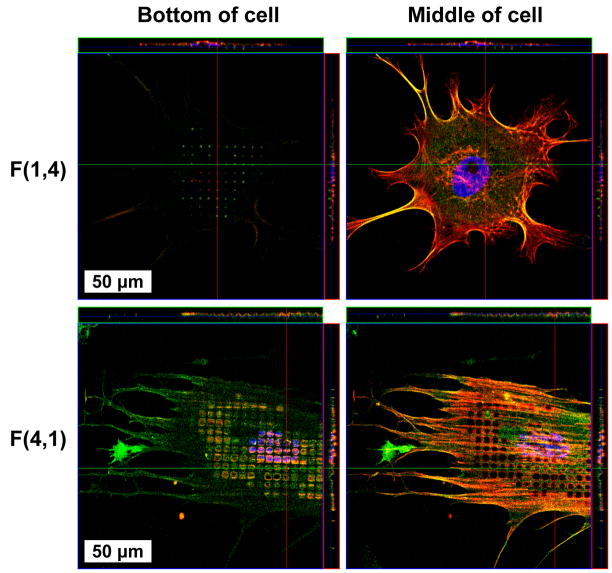
Cytoskeletal fusion proteins expressed in NHDFs were observed protruding 2.4 μm below the rest of the basal membrane plane from cells cultured on both 4 and 1 μm pits (Figure 6). Cells cultured on 4 μm pits appeared as “waves” in the XY and XZ planes, because both the apical and basal membrane descended into pits. This is in contrast to cells on 1 μm pits, which only extended their basal membrane surface to the pit bottom. Further, in many of the cells cultured on 1 μm pits, there were no protrusions visible.
Figure 6.
Confocal microscopy revealed NHDFs exploring the bottom of 1 and 4 μm diameter pits. The appearance of “holes” in the cell membrane of cells cultured on pattern F(4,1) is a consequence of the apical and basal cell surfaces bending below the focal plane, whereas cells on F(1,4) only reached the bottom of pits with their basal membrane.
4. Discussion
In this study, our goal was to test the hypothesis that topography, which has been shown to affect many cell phenotypes, also has an influence on cell transfectability. High-throughput microscopy of primary NHDFs cultured on topographical arrays revealed that cells on large, closely spaced pits were transfected a significant 25% more efficiently than those on smooth surfaces, while small, distant pits did not significantly alter GFP expression (Figure 2a). Flow cytometric analysis of NHDFs cultured on single patterns verified the prediction made by screening (Figure 4). To our knowledge, these results represent the first demonstration of a topographic control of transfection.
Additional experiments and measurements were performed in an effort to understand why cells cultured on patterned topography were transfected more efficiently. NHDFs that were least-spread were most efficiently transfected (Figures 2a, 2b, 3). Visible deformation of pattern F(4,1) but not of pattern F(1,4) implies cells may experience a different effective surface modulus and intracellular tension on each topography (Figure 5)– there is less contractile force required to deform pattern F(4,1). Cytoskeletal tension and cell spreading have been repeatedly demonstrated to modulate genome-wide changes in protein expression levels and patterns, including many involved in endocytosis: clathrin, epsin 2, caveolin-1, REP-2, and integrins [42–45]. Actin contraction, a process required for formation and strengthening of focal adhesions on integrin ligands during spreading, has been suggested to drive the internalization of cholesterol-rich lipid rafts containing cationic complexes bound to proteoglycans [46].
Emerging research suggests cell spreading may have a direct impact on endocytic processes. The formation, maturation, contraction, and disassembly of focal adhesions allow cells to spread over and move across their substrates. The speed of this cycle is controllable with substrate topography, as mediated through the heterogeneous spatial availability of adsorbed integrin ligands [15, 47]. A loss of integrin-mediated adhesion results in dramatic internalization of caveolae [34]. Disassembly of focal adhesions is a clathrin-dependent endocytic process; clathrin colocalizes with focal adhesions as nucleated by actin fibers, and knockdown of clathrin activity results in diminished integrin internalization [32]. These studies demonstrate strong links between integrin function and caveolae- and clathrin-mediated trafficking. These links could manifest as a coincident uptake of complexes or as a downregulation of particle uptake through competition for the endocytic machinery by altered integrin presentation and internalization on patterned topographies. As such, enhanced transfection efficiency of NHDFs cultured on patterned topography may be a downstream consequence of altered endocytic uptake and/or intracellular trafficking.
Substrate stiffness has been previously demonstrated to control transfectability, with stiffer substrates supporting more efficient transfection, putatively a consequence of enhanced proliferation on stiff surfaces [3]. Similarly, dense presentation of integrin ligands has also been shown to improve gene transfer through increased proliferation [48]. In our study however, neither pit spacing nor size had a significant impact on cell density or proliferation compared to smooth substrates (Figures 2c, 2d), indicating differences in cell division are not responsible for the enhanced transfection efficiency. Regions with large pit spacing had a greater number of cells than those with small spacing after transfection (Figure 2c); when taken with the observation that proliferation is unchanged this suggests either decreased initial attachment, or survival, during transfection on more discontinuous surfaces.
Luciferase expression levels were not detectably altered with transfection on the patterns that increased the percentage of GFP-expressing cells. This result may reflect the difference in how the reporter signals are weighted in each case - weakly GFP+ cells are counted with equal importance as those that saturate the photodetectors when quantified with thresholded fluorescence microscopy or flow cytometry. In contrast, luciferase assays weight highly-expressing cells more heavily than weakly-positive cells. Therefore, patterned topography may be exerting its influence on cells that express only a few copies of reporter - that is, on cells that are very near to the positive/negative threshold. Alternatively, luciferase expression could be more toxic than GFP in NHDFs, leading to greater cell death in luciferase-positive cells. Increased transfection efficiency without an associated increase in transfection level is desirable at times, particularly for transfected cell arrays, where over-expression can be a significant problem if it doesn’t reflect a natural phenotype in vivo [49].
Detailed microscopic examination of cells cultured on patterned topography reveals more possibilities to explain the increase in transfection efficiency. SEM images demonstrate NHDFs stretch their membrane over open pits as they move across the substrate, thereby exposing their basal surface to complex-containing medium (Figure 5). It is reasonable to expect this process to occur more often, and for a larger proportion of the cell membrane, on surfaces with large, dense pits. Including sidewall surfaces, dense pits have an increased effective surface area than sparse patterns or smooth surfaces; pattern F(4,1) has ~ 2.4 and 1.8× greater effective surface area than smooth and F(1,4) substrates, respectively. This potentially increases the amount of fibronectin that is able to adsorb to pitted surfaces, and the amount of nonviral vector that can adsorb during transfection. Substrate-mediated gene delivery has at times been demonstrated to be more effective than bolus delivery [4, 6, 50]; if cells can interact with this increased substrate surface area, they may have access to a larger amount of complexes, increasing their probability of being successfully transfected. Confocal microscopy revealed that NHDFs were indeed capable of reaching the bottom of both 1 and 4 μm pits (Figure 6), though the cells were more able to do so for 4 μm pits, which may have contributed to their enhanced transfection.
Cell culture substrates for in vitro use are typically smooth by default, whereas implantable gene-eluting scaffolds often impart tunable porosity (and therefore topography) via electrospinning [51], polymer chemistry, or porogen inclusion [52] to promote cell penetration and enhance mass transfer of entrapped therapeutics. The microenvironment at implant surfaces is critically important to efficient transfection; the inclusion of soluble factors, adsorption of ECM molecules, maintenance of cell-cell interactions, and tuning of mechanical stress are all currently leveraged towards the improvement of transfection efficiency [53]. Scaffold surface topography is an additional design parameter that may be optimized to bolster in situ genetic manipulation of infiltrating cells.
Ultimately, our goal is to understand the functional relationship between substrate topography, focal adhesion formation, cytoskeletal tension, endocytosis, and transfectability. We elected to take the first step with demonstration of the most downstream effect, transfectability, using bolus delivery of ubiquitous Lipofectamine 2000 complexes to primary human fibroblasts cultured on a combinatorial library of well-defined pitted topographies. Transfection is a multi-step process with many known and unknown modulators, with some that are evidently tunable with substrate topography. Discovery of topographies that increase transfection efficiency in primary human fibroblasts is another example of the power of high-throughput screening, complimenting the previous use of this technique to discover substrates with desirable stem and progenitor cell maintenance [54] or differentiation [39] effects and cytoskeletal changes in primary fibroblasts [55]. Our future work is focused on the extension of this finding to nanotopographic surfaces for substrate-mediated transfection, which we hypothesize to be even more potent than microtopographic influences on bolus transfection.
5. Conclusions
We have performed a systematic screen of nonviral transfection on microtopographic substrates, and discovered a significant 25% enhancement of transfection efficiency for primary human fibroblasts cultured on densely-pitted surfaces compared to smooth. This improvement could not be attributed to altered proliferation rate for cells on pitted substrates, and represents the first demonstration of a topographic influence on nonviral gene transfer. Topographic modulation of transfection is a new and therefore under-appreciated factor to consider when developing and studying gene delivery systems.
Supplementary Material
Supplementary Figure 1. An example of BrdU incorporation and staining in NHDFs following transfection. For illustration, BrdU-positive and DAPI-positive nuclei are false-colored green and red, respectively. Nuclei that have undergone division are double-positive, and appear yellow.
Supplementary Figure 2. High-resolution composite images (4× objective) of GFP expression and actin and nuclei staining on topographical arrays were overlaid with masks delineating each patterned region, and each region was analyzed sequentially with ImageJ.
Supplementary Figure 3. Examples of flow cytometric quantification of transfection efficiency in NHDFs cultured on F(4,1), F(1,4), and smooth PDMS. Dead cells were excluded from analysis with gates on forward- and side-scatter (left panel) and propidium iodide (right panel). The broad propidium iodide peak is a result of complexes bound to cell surfaces. Bright population of GFP-expressing cells saturated the photodetector (center panel). Untransfected cells cultured on smooth PDMS served as negative controls (smooth NC), with 1% gated as GFP+.
Supplementary Figure 4. Freeze-fracture SEM of patterns F(1,4) and F(4,1) revealed a uniform depth of 2.4 μm.
Acknowledgments
Support by NIH (HL83008), AOSpine International, and the Center for Biomolecular and Tissue Engineering (CBTE, Duke University) NIH Training Program is acknowledged. We are also grateful for: α-actinin-GFP plasmid from Dr. Carol A. Otey (University of North Carolina at Chapel Hill, Chapel Hill, NC), FUGW plasmid from Dr. David Baltimore (California Institute of Technology, Pasadena, CA), psPAX2 and pMD2.G plasmids from Dr. Didier Trono (Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland), and Lifeact-mRFPRuby plasmid, a generous gift from Dr. Roland Wedlich-Söldner (Max Planck Institute of Biochemistry, Martinsried, Germany).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adler AF, Leong KW. Emerging links between surface nanotechnology and endocytosis: impact on nonviral gene delivery. Nano Today. 2010;5(6):553–69. doi: 10.1016/j.nantod.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blacklock J, Vetter A, Lankenau A, Oupický D, Möhwald H. Tuning the mechanical properties of bioreducible multilayer films for improved cell adhesion and transfection activity. Biomaterials. 2010;31(27):7167–74. doi: 10.1016/j.biomaterials.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nat Mater. 2005;4(6):460–4. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 4.Yoshikawa T, Uchimura E, Kishi M, Funeriu DP, Miyake M, Miyake J. Transfection microarray of human mesenchymal stem cells and on-chip siRNA gene knockdown. J Control Release. 2004;96(2):227–32. doi: 10.1016/j.jconrel.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Uchimura E, Yamada S, Nomura T, Matsumoto K, Fujita S, Miyake M, et al. Reverse transfection using antibodies against a cell surface antigen in mammalian adherent cell lines. J Biosci Bioeng. 2007;104(2):152–5. doi: 10.1263/jbb.104.152. [DOI] [PubMed] [Google Scholar]
- 6.Bengali Z, Pannier AK, Segura T, Anderson BC, Jang JH, Mustoe TA, et al. Gene delivery through cell culture substrate adsorbed DNA complexes. Biotechnol Bioeng. 2005;90(3):290–302. doi: 10.1002/bit.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pannier AK, Anderson BC, Shea LD. Substrate-mediated delivery from self-assembled monolayers: effect of surface ionization, hydrophilicity, and patterning. Acta Biomater. 2005;1(5):511–22. doi: 10.1016/j.actbio.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pannier AK, Wieland JA, Shea LD. Surface polyethylene glycol enhances substrate-mediated gene delivery by nonspecifically immobilized complexes. Acta Biomater. 2008;4(1):26–39. doi: 10.1016/j.actbio.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem. 2002;13(3):621–9. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 10.Mehrotra S, Lee I, Chan C. Multilayer mediated forward and patterned siRNA transfection using linear-PEI at extended N/P ratios. Acta Biomater. 2009;5(5):1474–88. doi: 10.1016/j.actbio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchimura E, Yamada S, Uebersax L, Fujita S, Miyake M, Miyake J. Method for reverse transfection using gold colloid as a nano-scaffold. J Biosci Bioeng. 2007;103(1):101–3. doi: 10.1263/jbb.103.101. [DOI] [PubMed] [Google Scholar]
- 12.Dhaliwal A, Maldonado M, Han Z, Segura T. Differential uptake of DNA-poly(ethylenimine) polyplexes in cells cultured on collagen and fibronectin surfaces. Acta Biomater. 2010;6(9):3436–47. doi: 10.1016/j.actbio.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bengali ZC, Rea J, Shea LD. Gene expression and internalization following vector adsorption to immobilized proteins: dependence on protein identity and density. J Gene Med. 2007;9(8):668–78. doi: 10.1002/jgm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medina-Kauwe LK, Xie J, Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther. 2005;12(24):1734–51. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- 15.Biggs MJP, Richards RG, Dalby MJ. Nanotopographical modification: a regulator of cellular function through focal adhesions. Nanomed-Nanotechnol. 2010;6(5):619–33. doi: 10.1016/j.nano.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lord MS, Foss M, Besenbacher F. Influence of nanoscale surface topography on protein adsorption and cellular response. Nano Today. 2010;5(1):66–78. [Google Scholar]
- 17.Carpenter J, Khang D, Webster TJ. Nanometer polymer surface features: the influence on surface energy, protein adsorption and endothelial cell adhesion. Nanotechnology. 2008;19(50):505103. doi: 10.1088/0957-4484/19/50/505103. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Garcia C, Sousa SR, Moratal D, Rico P, Salmeron-Sanchez M. Effect of nanoscale topography on fibronectin adsorption, focal adhesion size and matrix organisation. Colloid Surface B. 2010;77(2):181–90. doi: 10.1016/j.colsurfb.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Hovgaard MB, Rechendorff K, Chevallier J, Foss M, Besenbacher F. Fibronectin adsorption on tantalum: the influence of nanoroughness. J Phys Chem B. 2008;112(28):8241–49. doi: 10.1021/jp801103n. [DOI] [PubMed] [Google Scholar]
- 20.Lord MS, Cousins BG, Doherty PJ, Whitelock JM, Simmons A, Williams RL, et al. The effect of silica nanoparticulate coatings on serum protein adsorption and cellular response. Biomaterials. 2006;27(28):4856–62. doi: 10.1016/j.biomaterials.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 21.Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Use of nanotopography to study mechanotransduction in fibroblasts - methods and perspectives. Eur J Cell Biol. 2004;83(4):159–69. doi: 10.1078/0171-9335-00369. [DOI] [PubMed] [Google Scholar]
- 22.Chalut KJ, Chen S, Finan JD, Giacomelli MG, Guilak F, Leong KW, et al. Label-free, high-throughput measurements of dynamic changes in cell nuclei using angle-resolved low coherence interferometry. Biophys J. 2008;94(12):4948–56. doi: 10.1529/biophysj.107.124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chalut KJ, Kulangara K, Giacomelli MG, Wax A, Leong KW. Deformation of stem cell nuclei by nanotopographical cues. Soft Matter. 2010;6(8):1675–81. doi: 10.1039/B921206J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CDW, Curtis ASG. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2003;284(2):274–82. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 25.Le Beyec J, Xu R, Lee S-Y, Nelson CM, Rizki A, Alcaraz J, et al. Cell shape regulates global histone acetylation in human mammary epithelial cells. Experimental Cell Research. 2007;313(14):3066–75. doi: 10.1016/j.yexcr.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12(3):468–74. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 27.Hoekstra D, Rejman J, Wasungu L, Shi F, Zuhorn I. Gene delivery by cationic lipids: in and out of an endosome. Biochem Soc Trans. 2007;35(Pt 1):68–71. doi: 10.1042/BST0350068. [DOI] [PubMed] [Google Scholar]
- 28.Lindberg J, Fernandez MA, Ropp JD, Hamm-Alvarez SF. Nocodazole treatment of CV-1 cells enhances nuclear/perinuclear accumulation of lipid-DNA complexes and increases gene expression. Pharm Res. 2001;18(2):246–9. doi: 10.1023/a:1011001022570. [DOI] [PubMed] [Google Scholar]
- 29.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–63. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 30.Wiesner S, Lange A, Fassler R. Local call: from integrins to actin assembly. Trends Cell Biol. 2006;16(7):327–9. doi: 10.1016/j.tcb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Jeng RL, Welch MD. Cytoskeleton: Actin and endocytosis - no longer the weakest link. Curr Biol. 2001;11(17):R691–R694. doi: 10.1016/s0960-9822(01)00410-9. [DOI] [PubMed] [Google Scholar]
- 32.Chao WT, Kunz J. Focal adhesion disassembly requires clathrin-dependent endocytosis of integrins. FEBS Lett. 2009;583(8):1337–43. doi: 10.1016/j.febslet.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wary KK, Mariotti A, Zurzolo C, Giancotti FG. A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth. Cell. 1998;94(5):625–34. doi: 10.1016/s0092-8674(00)81604-9. [DOI] [PubMed] [Google Scholar]
- 34.del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RGW, Schwartz MA. Integrins regulate Rac targeting by internalization of membrane domains. Science. 2004;303(5659):839–42. doi: 10.1126/science.1092571. [DOI] [PubMed] [Google Scholar]
- 35.Echarri A, Del Pozo MA. Caveolae internalization regulates integrin-dependent signaling pathways. Cell Cycle. 2006;5(19):2179–82. doi: 10.4161/cc.5.19.3264. [DOI] [PubMed] [Google Scholar]
- 36.Martinez E, Engel E, Planell JA, Samitier J. Effects of artificial micro- and nano-structured surfaces on cell behaviour. Ann Anat. 2009;191(1):126–35. doi: 10.1016/j.aanat.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Yim EK, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials. 2005;26(26):5405–13. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berry CC, Campbell G, Spadiccino A, Robertson M, Curtis AS. The influence of microscale topography on fibroblast attachment and motility. Biomaterials. 2004;25(26):5781–8. doi: 10.1016/j.biomaterials.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 39.Lovmand J, Justesen J, Foss M, Lauridsen RH, Lovmand M, Modin C, et al. The use of combinatorial topographical libraries for the screening of enhanced osteogenic expression and mineralization. Biomaterials. 2009;30(11):2015–22. doi: 10.1016/j.biomaterials.2008.12.081. [DOI] [PubMed] [Google Scholar]
- 40.Riedl J, Flynn KC, Raducanu A, Gartner F, Beck G, Bosl M, et al. Lifeact mice for studying F-actin dynamics. Nat Meth. 2010;7(3):168–9. doi: 10.1038/nmeth0310-168. [DOI] [PubMed] [Google Scholar]
- 41.Edlund M, Lotano MA, Otey CA. Dynamics of alpha-actinin in focal adhesions and stress fibers visualized with alpha-actinin-green fluorescent protein. Cell Motil Cytoskel. 2001;48(3):190–200. doi: 10.1002/1097-0169(200103)48:3<190::AID-CM1008>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 42.Dalby MJ, Gadegaard N, Wilkinson CDW. The response of fibroblasts to hexagonal nanotopography fabricated by electron beam lithography. J Biomed Mater Res A. 2008;84A(4):973–9. doi: 10.1002/jbm.a.31409. [DOI] [PubMed] [Google Scholar]
- 43.Dalby MJ, Yarwood SJ, Johnstone HJ, Affrossman S, Riehle MO. Fibroblast signaling events in response to nanotopography: a gene array study. IEEE Trans Nanobioscience. 2002;1(1):12–17. doi: 10.1109/tnb.2002.806930. [DOI] [PubMed] [Google Scholar]
- 44.Dalby MJ, Yarwood SJ, Riehle MO, Johnstone HJH, Affrossman S, Curtsi ASG. Increasing fibroblast response to materials using nanotopography: morphological and genetic measurements of cell response to 13-nm-high polymer demixed islands. Exp Cell Res. 2002;276(1):1–9. doi: 10.1006/excr.2002.5498. [DOI] [PubMed] [Google Scholar]
- 45.Yim EK, Pang SW, Leong KW. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp Cell Res. 2007;313(9):1820–9. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopatz I, Remy J-S, Behr J-P. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J Gene Med. 2004;6(7):769–76. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 47.Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys J. 2007;92(8):2964–74. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong HJ, Hsiong S, Mooney DJ. Nanoscale cell adhesion ligand presentation regulates nonviral gene delivery and expression. Nano Lett. 2006;7(1):161–6. doi: 10.1021/nl062485g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey SN, Wu RZ, Sabatini DM. Applications of transfected cell microarrays in high-throughput drug discovery. Drug Discov Today. 2002;7(18 Suppl):S113–8. doi: 10.1016/s1359-6446(02)02386-3. [DOI] [PubMed] [Google Scholar]
- 50.Fujita S, Ota E, Sasaki C, Takano K, Miyake M, Miyake J. Highly efficient reverse transfection with siRNA in multiple wells of microtiter plates. J Biosci Bioeng. 2007;104(4):329–33. doi: 10.1263/jbb.104.329. [DOI] [PubMed] [Google Scholar]
- 51.Chakraborty S, Liao IC, Adler A, Leong KW. Electrohydrodynamics: a facile technique to fabricate drug delivery systems. Adv Drug Deliver Rev. 2009;61(12):1043–54. doi: 10.1016/j.addr.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y, De Laporte L, Rives CB, Jang JH, Lin WC, Shull KR, et al. Neurotrophin releasing single and multiple lumen nerve conduits. J Control Release. 2005;104(3):433–46. doi: 10.1016/j.jconrel.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Laporte L, Shea LD. Matrices and scaffolds for DNA delivery in tissue engineering. Adv Drug Deliver Rev. 2007;59(4–5):292–307. doi: 10.1016/j.addr.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markert LD, Lovmand J, Foss M, Lauridsen RH, Lovmand M, Fuchtbauer EM, et al. Identification of distinct topographical surface microstructures favoring either undifferentiated expansion or differentiation of murine embryonic stem cells. Stem Cells Dev. 2009;18(9):1331–42. doi: 10.1089/scd.2009.0114. [DOI] [PubMed] [Google Scholar]
- 55.Kolind K, Dolatshahi-Pirouz A, Lovmand J, Pedersen FS, Foss M, Besenbacher F. A combinatorial screening of human fibroblast responses on micro-structured surfaces. Biomaterials. 2010;31(35):9182–91. doi: 10.1016/j.biomaterials.2010.08.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. An example of BrdU incorporation and staining in NHDFs following transfection. For illustration, BrdU-positive and DAPI-positive nuclei are false-colored green and red, respectively. Nuclei that have undergone division are double-positive, and appear yellow.
Supplementary Figure 2. High-resolution composite images (4× objective) of GFP expression and actin and nuclei staining on topographical arrays were overlaid with masks delineating each patterned region, and each region was analyzed sequentially with ImageJ.
Supplementary Figure 3. Examples of flow cytometric quantification of transfection efficiency in NHDFs cultured on F(4,1), F(1,4), and smooth PDMS. Dead cells were excluded from analysis with gates on forward- and side-scatter (left panel) and propidium iodide (right panel). The broad propidium iodide peak is a result of complexes bound to cell surfaces. Bright population of GFP-expressing cells saturated the photodetector (center panel). Untransfected cells cultured on smooth PDMS served as negative controls (smooth NC), with 1% gated as GFP+.
Supplementary Figure 4. Freeze-fracture SEM of patterns F(1,4) and F(4,1) revealed a uniform depth of 2.4 μm.