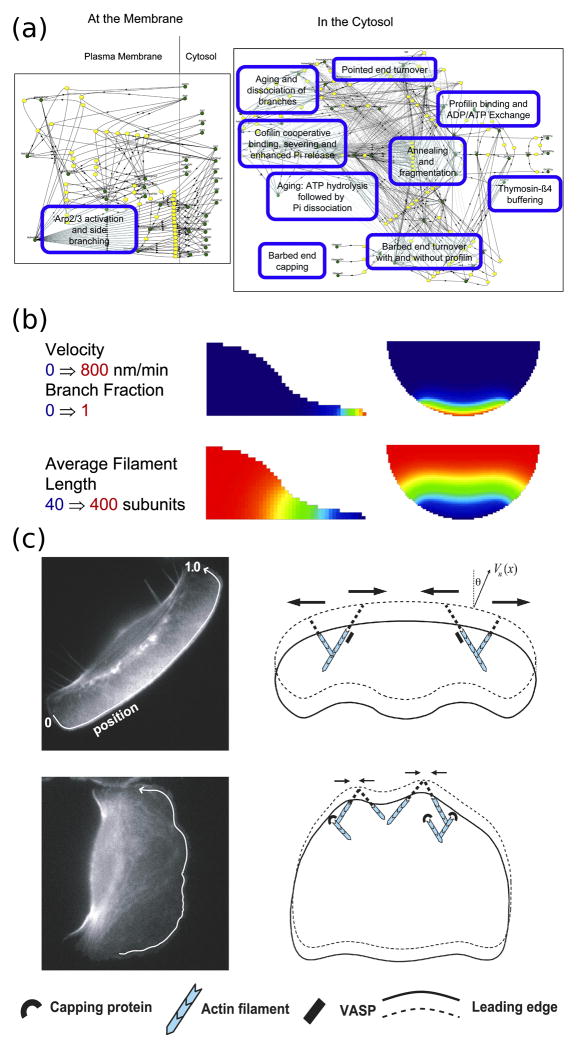
Figure 2. Computational modeling of actin cytoskeletal dynamics.
(a) The continuum model assembled by Ditlev et al. encompasses an extensive actin modification network. In the diagram, green circles represent different species or state variables, connected to the reactions (yellow) by solid lines, and the dashed lines represent catalytic interactions. (Reprinted with permission from [24]. Copyright 2009 Elsevier Inc.). (b) Implementation of the model by Ditlev et al. in three dimensions following activation of dendritic nucleation at the cell edge. The top row shows how the protrusion velocity (calculated as a function of actin branching) is localized to the cell front and decays rapidly behind the leading edge. The bottom row shows how actin filament length depends on the rapid nucleation and capping of actin filaments at the cell front that results from depletion of G-actin. (Reprinted with permission from [24]. Copyright 2009 Elsevier Inc.). (c) The model of actin filament dynamics by Lacayo et al. shows how excessive filament capping at the leading edge can produce rough cell morphologies. In cells with high VASP activity at the leading edge (top row), actin filaments are protected from capping and are subject to lateral flows that smooth heterogeneities in protrusive force at the leading edge. (Reprinted with permission from [26]).