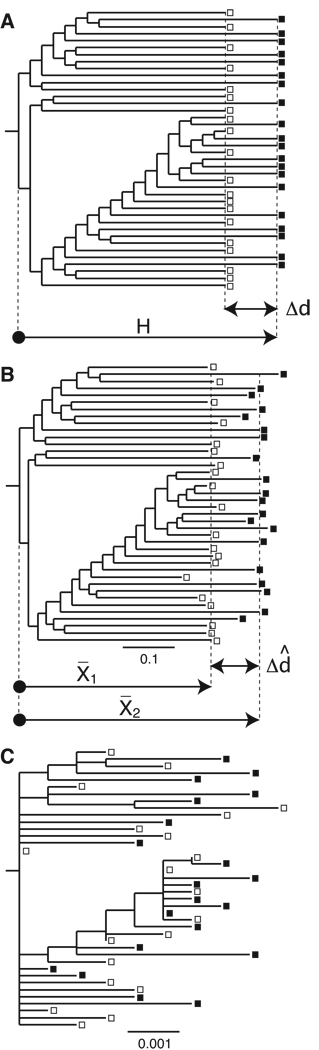
Figure 1. Definitions and examples of simulated trees.
(A) An example of a randomly generated true tree, with perfect “clocklike” edges. H is the total tree height, and Δd is the true (expected) distance between sample 1 and 2 OTUs. This tree is at Δd/H=0.2 and 20 OTUs in each sample. Thus, this tree shows the definitions of Δd and H, and is the true tree on which the trees in panels B and C were simulated, allowing for comparison between estimated rate and expected rate (Δd̂/Δd). (B) The same tree topology with Poisson distributed edges, and scaled so that Δd = 0.1 substitutions/site. X̅1 is the average distance from the root to sample 1 OTUs, X̅2 is the average distance from the root to sample 2 OTUs, and Δd̂ is the estimated rate between the samples. (C) The same tree topology with Poisson distributed edges, and scaled so that Δd = 0.001 substitutions/site. Note that many expected short edges become zero at this low rate, and samples 1 and 2 are not well separated. Open squares are sample 1 OTUs and filled squares sample 2 OTUs. Trees in B and C are examples of trees used in evaluating our method, scaled to the shown scale bars. The tree in A is of arbitrary length.