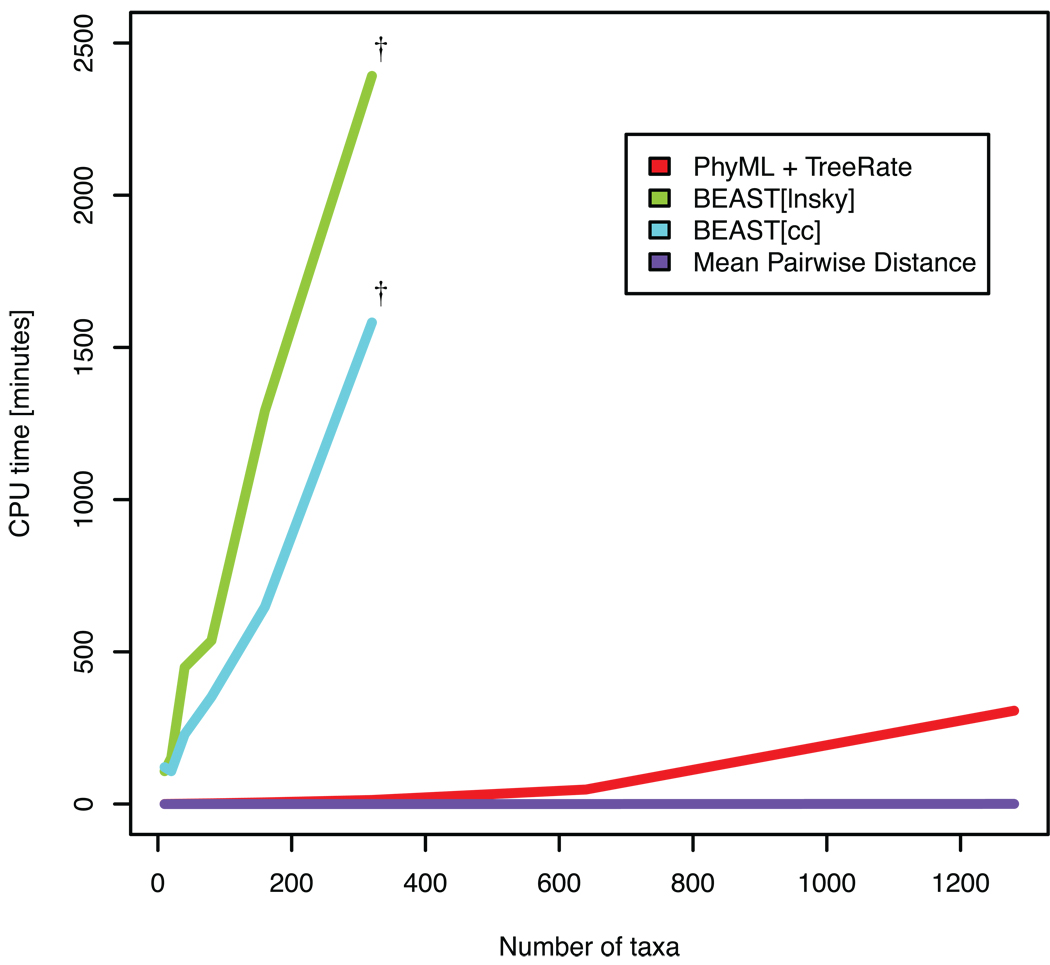
Figure 4. Runtime comparison to other methods.
TreeRate, including reconstruction of the tree using PhyML, calculation time was compared to two other methods that calculate evolutionary distances and rates, mean pariwise distance and BMCMC using two different population growth and molecular clock models (BEAST[cc] and BEAST[lnsky]), as described in Methods. The dagger symbol (†) indicates the last data size (320 taxa) that was possible to start using default settings in BEAST.