The first high-angle neutron fibre diffraction results from an amyloid system are described. These were obtained using H2O/D2O isototopic replacement, and emphasise the ability of neutron fibre diffraction, used in conjunction with deuterium labelling approaches, to study novel aspects of amyloid conformation and hydration.
Keywords: amyloid, neutron fibre diffraction, H2O/D2O isotopic replacement
Abstract
The first neutron fibre diffraction studies of an amyloid system are presented. The techniques used to prepare the large samples needed are described, as well as the procedures used to isotopically replace H2O in the sample by D2O. The results demonstrate the feasibility of this type of approach for the pursuit of novel structural analyses that will strongly complement X-ray fibre diffraction studies and probe aspects of amyloid structure that to date have remained obscure. The approach is demonstrated using an amyloid form of the peptide NSGAITIG, but is equally applicable for the study of other systems such as Alzheimer’s Aβ peptide.
1. Introduction
Amyloid and amyloid-type systems are being studied extensively because they are believed to be implicated in a wide variety of diseases such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, type 2 diabetes mellitus and a variety of transmissible spongiform encephalopathies (Pepys, 2001 ▶). While extensive structural studies have been carried out on a variety of different amyloid forms (Kirschner et al., 1986 ▶; Serpell, 2000 ▶; Sunde et al., 1997 ▶; Dobson, 2001 ▶; Jaroniec et al., 2004 ▶; Wille et al., 2009 ▶; Inouye et al., 2010 ▶; Paravastu et al., 2009 ▶), major questions remain about the amyloid deposits associated with these pathologies. Amyloids are typically characterized using Congo red staining, electron microscopy and X-ray fibre diffraction. Electron microscopy shows well defined filaments with diameters of approximately 100 Å and X-ray fibre diffraction work suggests that these consist of stacked β-sheets with the hydrogen-bonded β-strands aligned perpendicular to the filament axis (Makin & Serpell, 2005 ▶; Sachse et al., 2008 ▶; White et al., 2009 ▶). The quality of amyloid fibre diffraction data is usually poor in comparison with the high-quality crystalline or continuous fibre diffraction data used in the definitive analyses of other filamentous systems; the diffraction patterns often suffer from poor alignment and typically consist of complex mixtures of continuous and sampled diffraction. While this disorder is likely to be of importance in itself, it makes diffraction analysis difficult. Consequently, several rather different models for amyloid fibrils have been proposed (Jahn et al., 2010 ▶). There is therefore a general feeling that aspects of the underlying motif of amyloid remain that are either absent from current models or at best poorly understood.
The strong interest in amyloid conformation has led to numerous studies of synthetic peptides that form similar filamentous cross-β conformations. The motivation for this has arisen in many cases from a desire to acquire better defined data that are more amenable to structure determination and in others from a specific biological context associated with the peptide sequences in question. For example, a seven-residue peptide from Sup35 has been studied in single crystals, yielding a structure in which the β-sheets are arranged in pairs held together by a water-free ‘steric zipper’, with successive β-strands linked by main-chain and side-chain hydrogen bonding (Nelson et al., 2005 ▶). Sawaya and coworkers subsequently reported similar types of structures for other amyloid-forming sequences and noted an extensive range of structures that can be related to amyloid fibrils (Sawaya et al., 2007 ▶). Another example is work that is being carried out on synthetic peptides with sequences derived from the shaft structure of adenovirus. This structure, which in its normal context is involved in docking between virus and host, exists as a highly stable triple β-spiral (van Raaij et al., 1999 ▶). It is believed to be held in this conformation by the C-terminal globular domain, which plays a ‘registration’ role; this role can also be played by a heterologous domain such as the fibritin ‘foldon’ (Papanikolopoulou, Teixeira et al., 2004 ▶). However, outside the viral context and in the absence of any such registration signal a number of these peptides form amyloid fibrils (Papanikolopoulou et al., 2005 ▶; Kasotakis et al., 2009 ▶; Papanikolopoulou, Forge et al., 2004 ▶).
The current work was designed to test the feasibility of using neutron fibre diffraction methods to probe aspects of amyloid structure that have proved impossible to study using X-rays and where valuable information may be available that could allow the various models to be evaluated in completely different ways. Of particular interest was the possible exploitation of H2O/D2O isotopic replacement in analysing the role of water. The use of neutron fibre diffraction in this way was pioneered in early studies of ordered water in polymeric DNA (Forsyth et al., 1989 ▶; Langan et al., 1992 ▶; Shotton et al., 1997 ▶). The peptide used in this study was the eight-amino-acid sequence NSGAITIG, which contains the last loop–strand sequence of the adenovirus shaft structure.
2. Sample preparation
Samples were prepared by dissolving the NSGAITIG peptide to a concentration of 10 mg ml−1 in distilled water. Individual aligned fibres with diameters of approximately 50 µm were then drawn from the solution using methods that have been described previously (Papanikolopoulou et al., 2005 ▶). These samples were screened for quality using X-ray diffraction. A large sample suitable for neutron diffraction was then constructed by mounting up to 150 fibres (corresponding to a total peptide mass of approximately 15 mg) in a parallel array that matched the diameter of the neutron beam. A photograph of the sample is shown in Fig. 1 ▶. Depending on the nature of the subsequent neutron experiments, the sample was stored in either an H2O- or a D2O-humidified atmosphere prior to diffraction work. During data collection, purpose-designed humidity-control systems were used to control the water content of the sample environment. Similar techniques have been used to study structural transitions or to optimize sample crystallinity in fibrous samples (Mahendrasingam et al., 1986 ▶; Forsyth et al., 1986 ▶; Pope et al., 1998 ▶; McDonald et al., 2008 ▶).
Figure 1.
Fibre array constructed for neutron diffraction experiments on the peptide NSGAITIG. The sample consists of approximately 150 individual fibres each having a diameter of 50 µm.
3. Neutron diffraction
Neutron fibre diffraction data were recorded using the D19 and D22 diffractometers at the Institut Laue–Langevin (ILL) for the high- and low-angle parts of the data collection, respectively. The details and scope of these instruments are described in general terms by Teixeira, Zaccai et al. (2008 ▶), Forsyth, Mason et al. (2001 ▶) and Mason et al. (in preparation). The new D19 diffractometer has been used in numerous neutron fibre diffraction studies of synthetic and biological polymers (Gardner et al., 2004 ▶; Nishiyama et al., 2008 ▶), for a wide range of problems in structural chemistry (Ahrens et al., 2001 ▶; Broder et al., 2002 ▶) and, more recently, for neutron protein crystallography (Kovalevsky et al., 2010 ▶). Dewhurst (2010 ▶) describes D22 in detail. For this study, D19 was used with a wavelength of 2.4 Å. D22 was used with a wavelength of 6 Å, a sample-to-detector distance of 2 m and a wavelength bandpass of 10% (Δλ/λ). For both sets of experiments the sample was mounted in a purpose-designed humidity chamber that allowed the relative humidity of the sample environment to be accurately controlled (Shotton et al., 1998 ▶). More recently a ‘humidity stream’ system described by Sanchez-Weatherby et al. (2009 ▶) has been used to carry out studies of phase changes and structural transitions in macromolecular systems (Russi et al., 2011 ▶).
4. X-ray diffraction
X-ray diffraction experiments were carried out on instruments ID14-1 (Wakatsuki et al., 1998 ▶), using a wavelength of 0.933 Å, and ID02 (Narayanan et al., 2001 ▶), using a wavelength of 0.992 Å, for the high- and low-angle parts of the data collection, respectively. The environment of the sample was maintained at a high level of relative humidity: in the case of ID02 this was achieved through the use of a purpose-designed equipment (analogous to that described in §3) and in the case of ID14-1 the system described by Sanchez-Weatherby et al. (2009 ▶) was used.
5. Results
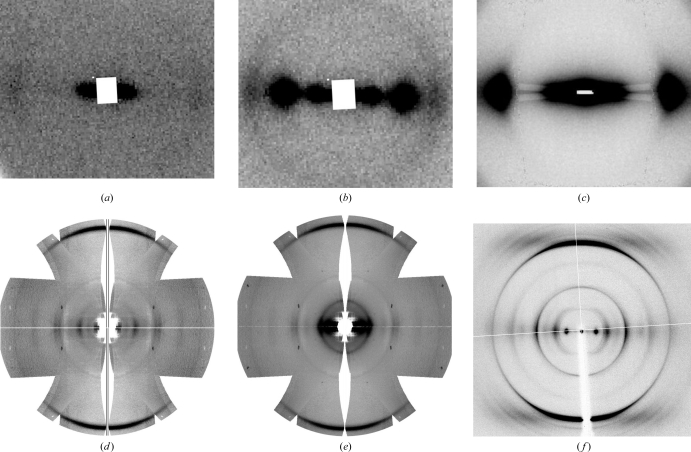
Fig. 2 ▶ shows the low- and high-angle neutron diffraction patterns recorded, alongside comparable X-ray diffraction patterns. The top row of images shows low-angle data extending to a high resolution limit of approximately 20 Å. The lower row (Figs. 2d–2f ▶) shows high-angle data to a resolution roughly corresponding to the 4.7 Å periodicity associated with the hydrogen-bonded β-strands of a cross-β-sheet. The change in the intensity distribution of the low-angle neutron diffraction data following H2O/D2O isotopic replacement is very clear from a comparison of Figs. 2 ▶(a) and 2 ▶(b). In the presence of H2O there is actually relatively little diffraction to be seen apart from a weak feature at approximately 50 Å. Following humidity-driven D2O replacement, two extremely strong peaks emerge at 50 and 25 Å. The high-angle neutron data seen in Figs. 2 ▶(d) and 2 ▶(e) also show significant intensity differences following H2O/D2O substitution, although the data are limited by counting statistics. Further data collection is planned to enhance the weaker parts of the diffraction pattern. For both low- and high-angle experiments tests were carried out to investigate the reproducibility and reversibility of the isotopic replacement. For example, separate experimental sessions were arranged during which both H2O and D2O data sets were measured but where in one case the sample had been pre-equilibrated in H2O and where in the other this had occurred in D2O. Hence, it was possible to be clear about the degree of isotopic exchange and to establish the reproducibility and reversibility of the effect.
Figure 2.
Neutron fibre diffraction data recorded from amyloid fibres of NSGAITIG in both H2O and D2O environments, shown alongside corresponding X-ray patterns. Low-angle data are shown in (a)–(c) and high-angle data in (d)–(f). (a, d) Neutron data from amyloid sample in H2O solvent. (b, e) Neutron data from amyloid sample in D2O solvent (c, f) X-ray data from amyloid sample in H2O solvent.
6. Discussion
This study demonstrates the feasibility of using neutron fibre diffraction methods to study amyloid systems. The sample preparation approach allows good sample orientation to be established and permits H2O/D2O replacement by humidification. While the idea of using neutron diffraction to study amyloid fibres was suggested in the 1970s (D. Kirschner, personal communication), we believe that this is the first account of such a study. It has been made possible as a result of major developments that have occurred for the D19 and D22 instruments at the ILL.
The results show that H2O/D2O substitution has profound effects on the observed neutron data and that the changes are fully reproducible and reversible. During the course of this work it was possible to assess the degree to which replacement of H2O by D2O might itself cause structural change to the amyloid fibrils. This was performed by recording X-ray fibre diffraction patterns from equivalently humidified fibres in H2O and D2O. The diffraction patterns recorded were essentially identical, indicating that within the resolution limits of the data H2O/D2O replacement was closely isomorphous. It is, therefore, reasonable to assume that both data sets could be used directly in modelling the distribution of water and exchangeable hydrogen within the amyloid system. This approach extends the scope of fibre diffraction methods for the study of amyloids and can be expected to be important in evaluating the different models that have been proposed for amyloids, including the role of hydration. The remaining stages of data collection and processing will shortly be completed; these are focused on improving the statistics for the high-angle data collected on D19 and also on processing a detailed low-angle contrast series covering the range from pure H2O to pure D2O solvent in steps of 10%. Given that (as in the case of comparable X-ray patterns) most of the accessible data are equatorial, initial modelling will focus on hydration as seen in projection along the fibril axis. At low resolution, simple geometric models will be considered for the location of water relative to peptide in the overall structure. This analysis will be extended to higher resolution using the available D19 data, yielding information on the distribution of ordered solvent and labile hydrogen.
The current work emphasizes the use of H2O/D2O substitution in amyloids, exploiting the large difference in the neutron scattering power of hydrogen and deuterium. More sophisticated experiments are possible through the use of perdeuterated or selectively deuterated samples. For example, analogous neutron fibre work on hydrogenated and perdeuterated analogues of the Alzheimer’s Aβ peptide and synuclein has begun.
The exploitation of deuteration methods is now being applied extensively in neutron crystallography (Teixeira, Blakeley et al., 2008 ▶; Blakeley, Langan et al., 2008 ▶; Blakeley, Ruiz et al., 2008 ▶), solution scattering (Laux et al., 2008 ▶; Vijayakrishnan et al., 2010 ▶; Callow et al., 2007 ▶) and dynamics (Jasnin et al., 2008 ▶). Such studies have only become possible since the inception of the Deuteration Laboratory at the ILL (Forsyth, Myles et al., 2001 ▶). Extensive opportunities exist for the application of these methods for the study of partially ordered systems; in the case of work on amyloids, they may be useful in understanding important aspects of structure and assembly.
Acknowledgments
SMT was supported by a Fulbright fellowship during the period of her stay at the Deuteration Laboratory within the Partnership for Structural Biology (PSB) at the Institut Laue–Langevin. EM was supported through a studentship held at Keele University with EU funding under contract STRP 033256. SC is grateful for a Keele University studentship. VTF acknowledges EPSRC support for the construction of the D19 diffractometer under grant GR/R47950/01 to Durham, Keele and Bath Universitues. VTF also acknowledges EPSRC support under grant EP/C015452/1 and from the EU under contract RII3-CT-2003-505925. We acknowledge the ILL for provision of neutron beamtime and the ESRF for time on beamlines ID02 and ID14.1.
References
- Ahrens, B., Davidson, M. G., Forsyth, V. T., Mahon, M. F., Johnson, A. L., Mason, S. A., Price, R. D. & Raithby, P. R. (2001). J. Am. Chem. Soc. 123, 9164–9165. [DOI] [PubMed]
- Blakeley, M. P., Langan, P., Niimura, N. & Podjarny, A. (2008). Curr. Opin. Struct. Biol. 18, 593–600. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Ruiz, F., Cachau, R., Hazemann, I., Meilleur, F., Mitschler, A., Ginell, S., Afonine, P., Ventura, O. N., Cousido-Siah, A., Haertlein, M., Joachimiak, A., Myles, D. & Podjarny, A. (2008). Proc. Natl Acad. Sci. USA, 105, 1844–1848. [DOI] [PMC free article] [PubMed]
- Broder, C. K., Davidson, M. G., Forsyth, V. T., Howard, J. A. K., Lamb, S. & Mason, S. A. (2002). Cryst. Growth Des. 2, 163–169.
- Callow, P., Sukhodub, A., Taylor, J. E. & Kneale, G. G. (2007). J. Mol. Biol. 369, 177–185. [DOI] [PMC free article] [PubMed]
- Dewhurst, C. (2010). ILL: Neutrons for Science: D22 http://www.ill.eu/instruments-support/instruments-groups/instruments/d22/.
- Dobson, C. M. (2001). Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 133–145. [DOI] [PMC free article] [PubMed]
- Forsyth, V. T., Greenall, R. J., Hussain, R., Mahendrasingam, A., Nave, C., Pigram, W. J. & Fuller, W. (1986). Biochem. Soc. Trans. 14, 553–557. [DOI] [PubMed]
- Forsyth, V. T., Mahendrasingam, A., Pigram, W. J., Greenall, R. J., Bellamy, K., Fuller, W. & Mason, S. A. (1989). Int. J. Biol. Macromol. 11, 236–240. [DOI] [PubMed]
- Forsyth, V. T., Mason, S. A., Howard, J. A. K., Davidson, M. G. & Fuller, W. (2001). Neutron News, 12(4), 20–25.
- Forsyth, V. T., Myles, D., Timmins, P. A. & Haertlein, M. (2001). Opportunities for Neutron Scattering in the 3rd Millennium, edited by J. Dianoux, pp. 47–54. Grenoble: Institut Laue–Langevin.
- Gardner, K. H., English, A. D. & Forsyth, V. T. (2004). Macromolecules, 37, 9654–9656.
- Inouye, H., Gleason, K. A., Zhang, D., Decatur, S. M. & Kirschner, D. A. (2010). Proteins, 78, 2306–2321. [DOI] [PubMed]
- Jahn, T. R., Makin, O. S., Morris, K. L., Marshall, K. E., Tian, P., Sikorski, P. & Serpell, L. C. (2010). J. Mol. Biol. 395, 717–727. [DOI] [PubMed]
- Jaroniec, C. P., MacPhee, C. E., Bajaj, V. S., McMahon, M. T., Dobson, C. M. & Griffin, R. G. (2004). Proc. Natl Acad. Sci. USA, 101, 711–716. [DOI] [PMC free article] [PubMed]
- Jasnin, M., Moulin, M., Haertlein, M., Zaccai, G. & Tehei, M. (2008). Biophys. J. 95, 857–864. [DOI] [PMC free article] [PubMed]
- Kasotakis, E., Mossou, E., Adler-Abramovich, L., Mitchell, E. P., Forsyth, V. T., Gazit, E. & Mitraki, A. (2009). Biopolymers, 92, 164–172. [DOI] [PubMed]
- Kirschner, D. A., Abraham, C. & Selkoe, D. J. (1986). Proc. Natl Acad. Sci. USA, 83, 503–507. [DOI] [PMC free article] [PubMed]
- Kovalevsky, A. Y., Hanson, L., Fisher, S. Z., Mustyakimov, M., Mason, S. A., Forsyth, V. T., Blakeley, M. P., Keene, D., Wagner, T., Carrell, H. L., Katz, A. K., Glusker, J. P. & Langan, P. (2010). Structure, 18, 688–699. [DOI] [PMC free article] [PubMed]
- Langan, P., Forsyth, V. T., Mahendrasingam, A., Pigram, W. J., Mason, S. A. & Fuller, W. (1992). J. Biomol. Struct. Dyn. 10, 489–503. [DOI] [PubMed]
- Laux, V., Callow, P., Svergun, D. I., Timmins, P. A., Forsyth, V. T. & Haertlein, M. (2008). Eur. Biophys. J. 37, 815–822. [DOI] [PubMed]
- Mahendrasingam, A., Forsyth, V. T., Hussain, R., Greenall, R. J., Pigram, W. J. & Fuller, W. (1986). Science, 233, 195–197. [DOI] [PubMed]
- Makin, O. S. & Serpell, L. C. (2005). FEBS J. 272, 5950–5961. [DOI] [PubMed]
- McDonald, M., Kendall, A., Tanaka, M., Weissman, J. S. & Stubbs, G. (2008). J. Appl. Cryst. 41, 206–209.
- Narayanan, T., Diat, O. & Boesecke, P. (2001). Nucl. Instrum. Methods Phys. Res. A, 467–468, 1005–1009.
- Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. Ø., Riekel, C., Grothe, R. & Eisenberg, D. (2005). Nature (London), 435, 773–778. [DOI] [PMC free article] [PubMed]
- Nishiyama, Y., Johnson, G. P., French, A. D., Forsyth, V. T. & Langan, P. (2008). Biomacromolecules, 9, 3133–3140. [DOI] [PubMed]
- Papanikolopoulou, K., Forge, V., Goeltz, P. & Mitraki, A. (2004). J. Biol. Chem. 279, 8991–8998. [DOI] [PubMed]
- Papanikolopoulou, K., Schoehn, G., Forge, V., Forsyth, V. T., Riekel, C., Hernandez, J. F., Ruigrok, R. W. & Mitraki, A. (2005). J. Biol. Chem. 280, 2481–2490. [DOI] [PubMed]
- Papanikolopoulou, K., Teixeira, S., Belrhali, H., Forsyth, V. T., Mitraki, A. & van Raaij, M. J. (2004). J. Mol. Biol. 342, 219–227. [DOI] [PubMed]
- Paravastu, A. K., Qahwash, I., Leapman, R. D., Meredith, S. C. & Tycko, R. (2009). Proc. Natl Acad. Sci. USA, 106, 7443–7448. [DOI] [PMC free article] [PubMed]
- Pepys, M. B. (2001). Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 203–210. [DOI] [PMC free article] [PubMed]
- Pope, L. H., Shotton, M. W., Forsyth, T., Hughes, D. J., Denny, R. C. & Fuller, W. (1998). Biophys. Chem. 70, 161–172. [DOI] [PubMed]
- Raaij, M. J. van, Mitraki, A., Lavigne, G. & Cusack, S. (1999). Nature (London), 401, 935–938. [DOI] [PubMed]
- Russi, S., Juers, D. H., Sanchez-Weatherby, J., Pellegrini, E., Mossou, E., Forsyth, V. T., Huet, J., Gobbo, A., Felisaz, F., Moya, R., McSweeney, S., Cusack, S., Bowler, M. W. & Cipriani, F. (2011). In the press. [DOI] [PubMed]
- Sachse, C., Fandrich, M. & Grigorieff, N. (2008). Proc. Natl Acad. Sci. USA, 105, 7462–7466. [DOI] [PMC free article] [PubMed]
- Sanchez-Weatherby, J., Bowler, M. W., Huet, J., Gobbo, A., Felisaz, F., Lavault, B., Moya, R., Kadlec, J., Ravelli, R. B. G. & Cipriani, F. (2009). Acta Cryst. D65, 1237–1246. [DOI] [PubMed]
- Sawaya, M. R., Sambashivan, S., Nelson, R., Ivanova, M. I., Sievers, S. A., Apostol, M. I., Thompson, M. J., Balbirnie, M., Wiltzius, J. J., McFarlane, H. T., Madsen, A. Ø., Riekel, C. & Eisenberg, D. (2007). Nature (London), 447, 453–457. [DOI] [PubMed]
- Serpell, L. C. (2000). Biochim. Biophys. Acta, 1502, 16–30. [DOI] [PubMed]
- Shotton, M. W., Pope, L. H., Forsyth, V. T., Denny, R. C., Archer, J., Langan, P., Ye, H. & Boote, C. (1998). J. Appl. Cryst. 31, 758–766.
- Shotton, M. W., Pope, L. H., Forsyth, T., Langan, P., Denny, R. C., Giesen, U., Dauvergne, M. T. & Fuller, W. (1997). Biophys. Chem. 69, 85–96. [DOI] [PubMed]
- Sunde, M., Serpell, L. C., Bartlam, M., Fraser, P. E., Pepys, M. B. & Blake, C. C. (1997). J. Mol. Biol. 273, 729–739. [DOI] [PubMed]
- Teixeira, S. C. M., Blakeley, M. P., Leal, R. M. F., Mitchell, E. P. & Forsyth, V. T. (2008). Acta Cryst. F64, 378–381. [DOI] [PMC free article] [PubMed]
- Teixeira, S. C. M., Zaccai, G. et al. (2008). Chem. Phys. 345, 133–151.
- Vijayakrishnan, S., Kelly, S. M., Gilbert, R. J. C., Callow, P., Bhella, D., Forsyth, T., Lindsay, J. G. & Byron, O. (2010). J. Mol. Biol. 399, 71–93. [DOI] [PMC free article] [PubMed]
- Wakatsuki, S., Belrhali, H., Mitchell, E. P., Burmeister, W. P., McSweeney, S. M., Kahn, R., Bourgeois, D., Yao, M., Tomizaki, T. & Theveneau, P. (1998). J. Synchrotron Rad. 5, 215–221. [DOI] [PubMed]
- White, H. E., Hodgkinson, J. L., Jahn, T. R., Cohen-Krausz, S., Gosal, W. S., Müller, S., Orlova, E. V., Radford, S. E. & Saibil, H. R. (2009). J. Mol. Biol. 389, 48–57. [DOI] [PMC free article] [PubMed]
- Wille, H., Bian, W., McDonald, M., Kendall, A., Colby, D. W., Bloch, L., Ollesch, J., Borovinskiy, A. L., Cohen, F. E., Prusiner, S. B. & Stubbs, G. (2009). Proc. Natl Acad. Sci. USA, 106, 16990–16995. [DOI] [PMC free article] [PubMed]