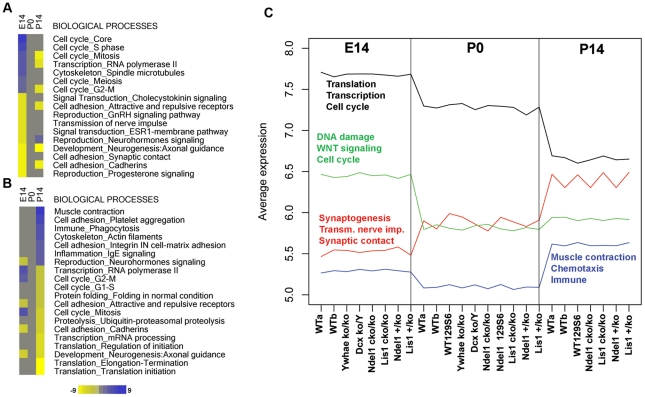
Figure 1. Cell cycle processes define early brain development in mouse, and unsupervised clustering revealed gene expression similarities in all genotypes at each developmental stage.
A,B) Differential expression analysis of WT brains followed by pathway analysis via Metacore software define biological processes that are up-/down-regulated at each developmental stage versus the other stages. A) Up-regulated (blue) and down-regulated processes (yellow) are sorted by intensity at E14. B) Similarly, up- and down-regulated processes are sorted by intensity at P14. No gene expression signature is characteristic of P0. Intensity values refer to enrichment values provided by Metacore and calculated as −Log(p-value) (FDR<0.05). C) Simultaneous analysis of gene expression data from all genotypes at all time points using the PAM algorithm, followed by gene enrichment (Metacore). PAM analysis revealed four patterns of gene expression that are similar across all genotypes, but different in a stage-dependent fashion. Black and green profiles represent genes highly expressed at E14, while red and blue represent genes with highest expression at P14. No particular expression pattern is characteristic of P0. Pathway analysis of the four patterns was consistent with the differential analysis results found in WT brains (see A,B).