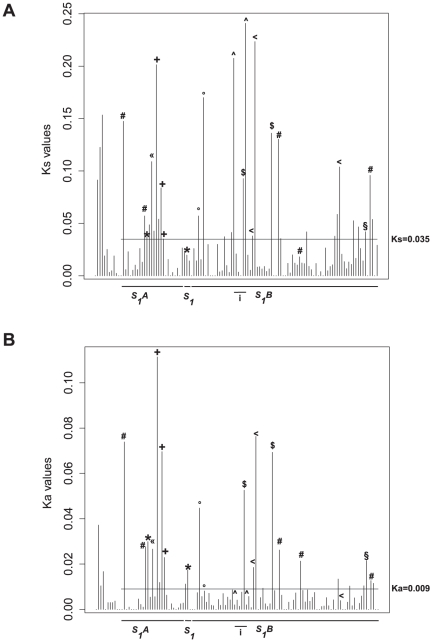
Figure 6. Representation of Ka and Ks values between 109 O. sativa and O. glaberrima orthologous genes in the S1 regions.
A. Representation of Ks values. The horizontal line is the mean value of Ks (Ks = 0.035) for the 109 analyzed orthologous genes in the S1 regions. B. Representation of Ka values. The horizontal line is the mean value of Ka (Ka = 0.09) for the 109 analyzed orthologous genes in the S1 regions. Lines below graphics represent analyzed genes located into S1A, S1, S1B loci, and those within the chromosomal inversion (i). Symbols represent different divergent genes with similar annotated function or unknown function as follows in O. glaberrima: (*) 66E18.4 and 49I08.11 (F-box proteins), (“) 66E18.6 (HAD phosphatase protein), (+) 66E18.8, 17A24.2 and 17A24.3 (LRR proteins), (°) 49I08.16 and 49I08.18 (Putative Serine/threonine protein kinases), (∧) 88O22.2 and 88O22.7 (Pectate lyase located at breakpoint inversion), (§) 41E07.23 (PRR protein) and (<) 88O22.10, 88O22.11 and 41E07.12 (Putative transferase proteins), ($) 88O22.6 and 88O22.19 (Hypothetical proteins) and (#) 93E08.17, 66E18.3, 56F23.3, 56F23.12 and 41E07.25 (Putative proteins).