Figure 1. Cross-species sequence conservation of miR-126, miR-150 and their target sites within the c-myb 3′UTR.
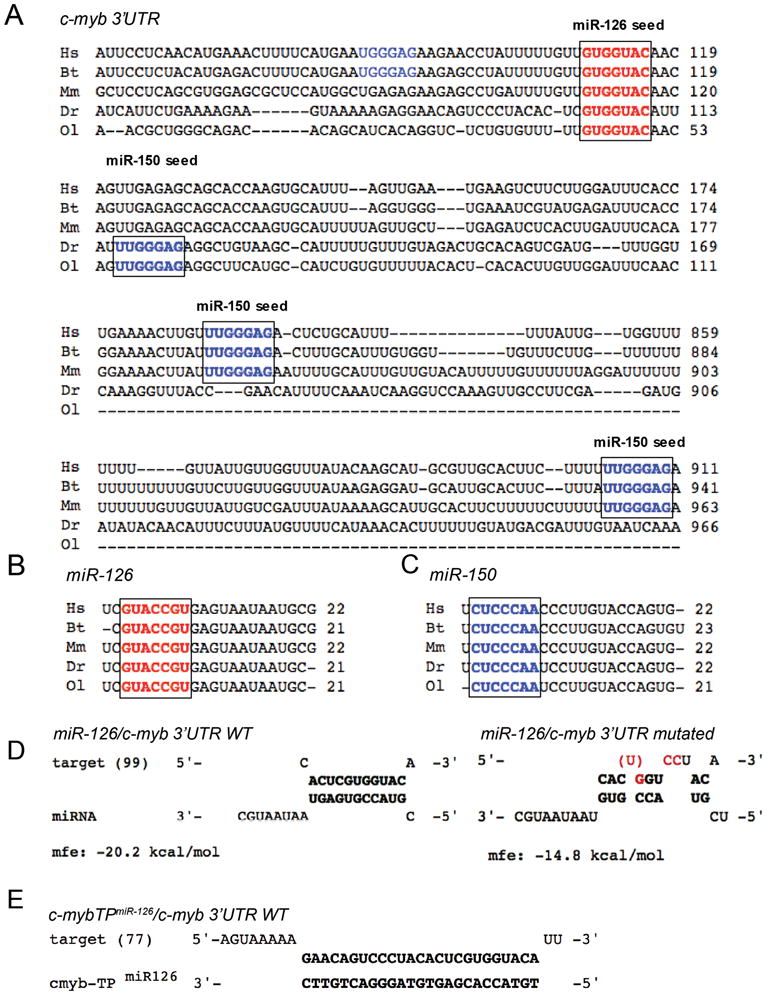
(A) 3′UTR sequences of c-myb flanking miR-126 (bold red) and mir-150 (bold and non-bold blue) target (‘seed’) sites were aligned among human (Hs), cattle (Bt), mouse (Mm), zebrafish (Dr) and medaka (Ol). Numbers indicate nucleotide positions following the STOP codon. For the single miR-126 site, nucleotide sequence and location within the 3′UTR are highly conserved among all species. For miR-150, two sites are highly conserved among human, cattle and mouse (bold blue); one additional site may be present in human and cattle (non-bold blue). Both fish 3′UTRs contain only one highly conserved miR-150 target site (bold blue). (B, C) The sequence of mature miR-126 (B) and miR-150 (C) is highly conserved among these species. The ‘seed’ sequence is highlighted (bold red) for miR-126 or (bold blue) for miR-150. UTR sequences were obtained from the ENSEMBL database (www.ensembl.org).
(D) We applied the web-based RNAhybrid tool to predict miRNA/mRNA interactions in zebrafish based on the minimum free energy hybridization of the wildtype (WT) or mutated c-myb 3′UTR and miR-126 54. The 5′-nucleotide position of the c-myb 3′UTR is indicated in brackets. Mutated or deleted nucleotides are highlighted in red. (E) To confirm that the putative miR-126 target site is used in vivo, the target protector morpholino c-myb-TPmiR126 was designed 100% complementary to the putative miR-126 target site within the zf-c-myb 3′UTR. If the predicted target site is used, competition of the c-myb-TPmiR126 morpholino and miR-126 for the same binding site will result in reduced or abolished regulation of c-myb by miR-126.
mfe, minimum free energy.
