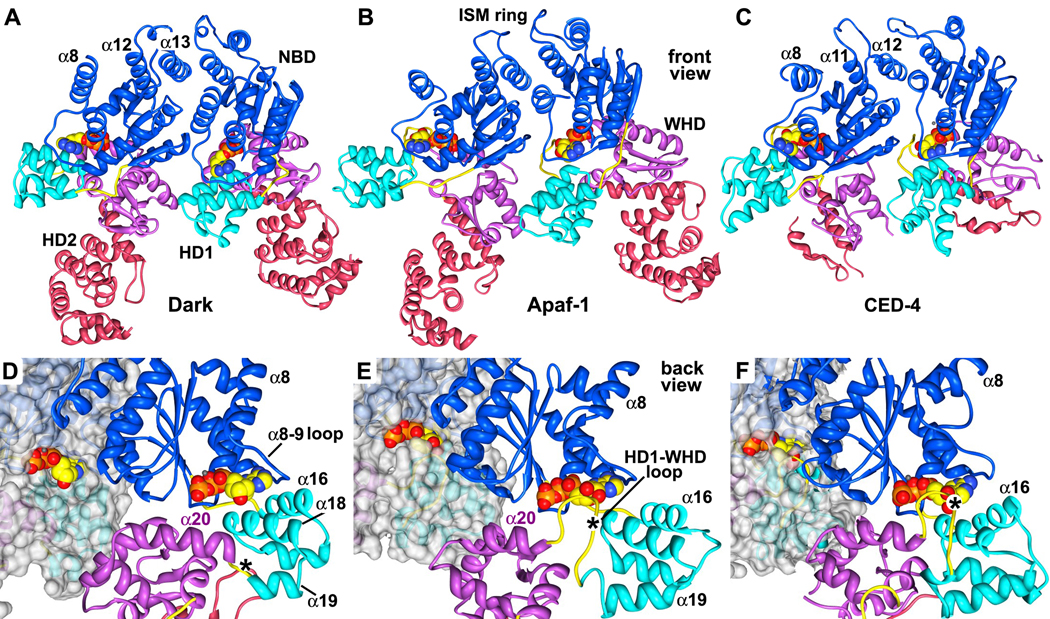
Figure 7. A comparison of lateral dimers and nucleotide binding sites in Dark, Apaf-1 and CED-4 apoptosomes.
A. A lateral dimer is shown from the Dark apoptosome. Structures in panels A–C have been aligned on the leftmost NBD.
B. A lateral dimer from the Apaf-1 apoptosome is shown. Differences in the packing of helix α8 are apparent.
C. The CED-4 lateral dimer is shown with its truncated HD2. The conformation of the α8 helix varies in the two subunits due to differences in packing of their respective CARDs in the disk (Qi et al., 2010).
D. A close-up is shown of the nucleotide binding region of Dark within the context of a lateral dimer. This view is from the bottom of the Dark single-ring. The subunit on the left has been rendered as a molecular surface to show the exposed nature of dATP. In panels D–F the lateral dimers are aligned on the rightmost NBD in each pair. The dATP binding pocket, as seen on the right, has a much shorter HD1-WHD loop than in the other apoptosomes (\marked with an asterisk).
E. A close-up of the same region in an Apaf-1 lateral dimer shows a more obscured path to bound ATP on the left, and the presence of a modeled HD1-WHD loop on the right, which embraces the nucleotide from the inward surface.
F. A similar close-up is shown for the atomic structure of the CED-4 dimer within the apoptosome. In this case, bound ATP is almost totally occluded within its binding pocket.
See also Figure S9.