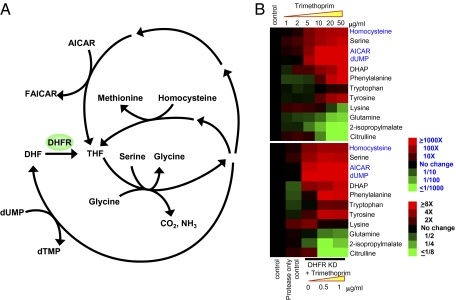
Fig. 5.
Metabolites affected by DHFR depletion or trimethoprim treatment. (A) Biochemical pathways related to DHFR. (B) Profiles of 12 intracellular metabolites in M. smegmatics treated with trimethoprim or depleted of DHFR. Relative levels are expressed as the log ratio of signal intensity normalized to the control sample, which was untreated cells. For dUMP, AICAR, and homocysteine, the numbers are in log10, and for the rest metabolites, the numbers are in log2. Both drug treatment and DHFR depletion were performed on the same strain, which has an ID-tagged DHFR and inducible protease on the chromosome. Protease-only control cells were induced for protease but not depleted for DHFR. Trimethoprim cells (B Upper) were treated with various concentrations of trimethoprim but not depleted for DHFR. DHFR-KD + trimethoprim (B Lower) indicates bacteria grown in the presence of various concentrations of trimethoprim and ATc 50 ng/mL, which depletes DHFR to less than 2% of its physiological concentration. This experiment was performed in duplicate, and this result is representative of three independent experiments. Metabolite signal intensities were normalized to total protein in the lysates.