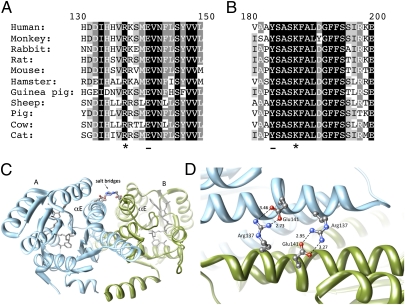
Fig. 2.
The structural importance of the 11β-HSD1 mutations R137C and K187N. (A) Alignment showing the strict conservation of Arg137 (marked with *) in 11β-HSD1 protein sequences from different species. Arg137 interacts to form a salt bridge with conserved residue Glu141 (underlined on the alignment) on the opposing subunit. (B) Alignment showing the conservation of Lys187 (marked with *) across species. Lys187 forms part of the catalytic tetrad, which includes the adjacent residue Tyr183 (underlined in the alignment). (C) A ribbons representation of the crystal structure of the 11β-HSD1 dimer (Protein Data Bank code: 2bel, chains A and B) showing the salt bridges between Arg137 and Glu141 on the αE helices of opposing subunits. Arg137 and Glu141 are represented in ball-stick format colored by heteroatom. (D) A closer view of the side-chain interactions between the two pairs of Arg137 and Glu141 residues at the subunit interface of 11β-HSD1 (Protein Data Bank code: 2bel, chains A and B), showing the close interaction (<4Å) between the heavy atoms. The salt bridges are of the fork-fork class (35) in this structure, although other 11β-HSD1 crystal structures are of the fork-stick type. These interactions may promote correct orientation of the subunits during dimer formation, as well as stabilize the final assembly. Structural images were produced using UCSF Chimera from the University of California San Francisco's Resource for Biocomputing, Visualization, and Informatics (supported by National Institutes of Health Grant P41 RR-01081) (36). Distances calculated between nitrogen and oxygen atoms are shown in Å.