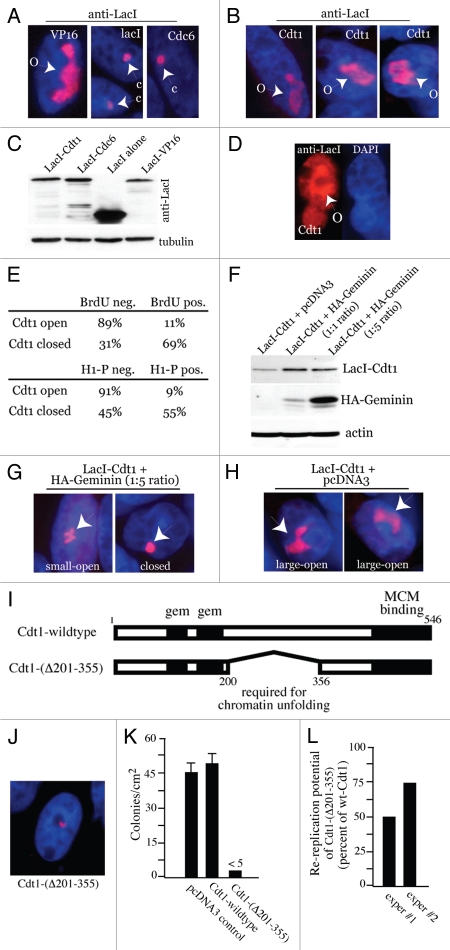
Figure 2.
Cdt1 targeting induces Geminin-sensitive large-scale chromatin decondensation in G1. A03_1 cells were used in (A–J). (A) LacI-VP16, LacI alone or LacI-Cdc6 were transiently expressed, followed by IF with anti-LacI and Texas Red to detect open/decondensed (‘O’) or closed/condensed (‘C’) HSRs. Nuclei are stained with DAPI. (B) LacI-Cdt1 was expressed and analyzed by IF to detect chromatin decondensation. (C) Immunoblot of LacI-fusion protein expression for the results in Table 1. (D) Anti-LacI IF separated from DAPI showing LacI-Cdt1 present throughout the nucleus. (E) LacI-Cdt1 was expressed for 24 h, then pulsed with BrdU. Anti-BrdU and anti-H1-P staining was used to relate the index of BrdU-negative and H1-P-positive cells to the open or closed HSR status. (F) HA-Geminin was transfected at a 5:1 or 1:1 plasmid ratio with LacI-Cdt1 and relative protein expression verified by IB. (G and H) Examples of small-open, closed and large-open HSRs for the indicated conditions. (I) Diagram showing location of Cdt1 chromatin unfolding domain. (J) Chromatin unfolding ability of Cdt1-(Δ201-355) was tested as above. (K) Colony forming assays were performed in CHO cells to test the ability of wt-Cdt1 and Cdt1-(4201-355) to suppress colony growth. Stable selection for protein expression lasted 14 days, followed by Giemsa staining. (L) HeLa cells were used as in Figure 1B to determine the re-replication ability of Cdt1-(Δ201-355) versus wt-Cdt1 except 48-h transient transfections were used. Results from two experiments are shown compared to wt-Cdt1 (normalized to 100% rereplication ability).