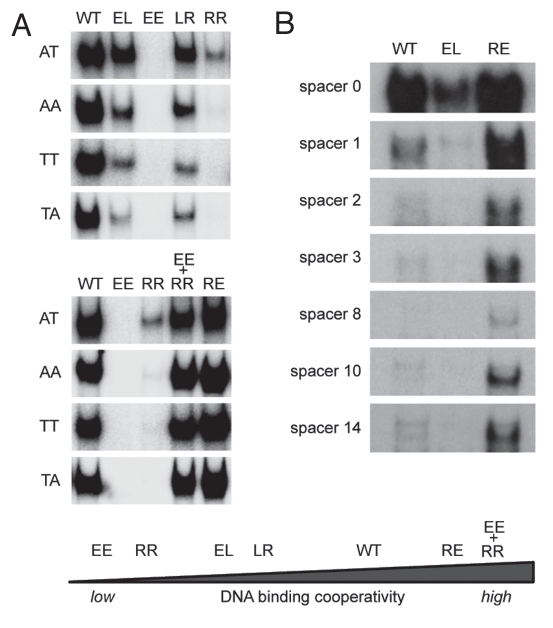
Figure 3.
Impact of DNA binding cooperativity on sequence selectivity of p53. (A) Shown are electrophoretic mobility shift assays (EMSA) for DNA binding of in vitro translated wild-type p53 and the indicated H1 helix mutants to dsDNA oligonucleotides (5′-GGG AGC TTA GGC WWG TCT AGG CWW GTC TA-3′) with WW denoting AT, AA, TT or TA sequences in the center of each half site. EMSAs were performed as previously described.9,72 Compared to H1 helix mutants with reduced DNA binding cooperativity (EE, RR, LR, EL), mutants with increased DNA binding cooperativity (RE and EE+RR) revealed an increased ability to bind the lower affinity non-CATG sequences. (B) Same as in (A) using dsDNA oligonucleotides containing the 5′ p53 binding site in the p21 promoter (5′-TCT GGC CGT CAG GAA CATG TCC (N)1–14 CAA CATG TTG AAG CTC TGG CAT A-3′) with increasing central spacer sequences (N)1–14. The high cooperativity mutant (RE) showed an increased ability to bind the spacer-containing motifs, while the low cooperativity mutant (EL) was largely unable to bind these spacer-containing elements.